| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:21:22 UTC |
|---|
| Updated at | 2022-12-13 23:36:26 UTC |
|---|
| CannabisDB ID | CDB006137 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Choline |
|---|
| Description | Choline, also known as bilineurine, is an organic compound containing a N, N, N-trimethylethanolammonium cation. Choline is an extremely weak basic (essentially neutral) compound (based on its pKa). Choline is a basic constituent of lecithin that is found in many plants and animal organs. It is important as a precursor of acetylcholine, as a methyl donor in various metabolic processes, and in lipid metabolism. In plants, such as Cannabis, the first step in de novo biosynthesis of choline involves the decarboxylation of serine into ethanolamine, which is catalyzed by a serine decarboxylase. The synthesis of choline from ethanolamine involves three consecutive N-methylation steps catalyzed by a methyl transferase. In terms of human health, choline is now considered to be an essential vitamin. While humans can synthesize small amounts (by converting phosphatidylethanolamine to phosphatidylcholine), it must be consumed in the diet to maintain health. Required levels are between 425 mg/day (female) and 550 mg/day (male). Milk, eggs, liver, and peanuts are especially rich in choline. Most choline is found in phospholipids, namely phosphatidylcholine or lecithin. Choline can be oxidized to form betaine, which is a methyl source for many reactions (i.e. conversion of homocysteine into methionine). Lack of sufficient amounts of choline in the diet can lead to fatty liver disease and general liver damage. This arises from the lack of VLDL , which is necessary to transport fats away from the liver. Choline deficiency also leads to elevated serum levels of alanine amino transferase and is associated with increased incidence of liver cancer. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Bilineurine | ChEBI | | CHOLINE ion | ChEBI | | N,N,N-Trimethylethanol-ammonium | ChEBI | | N-Trimethylethanolamine | ChEBI | | Trimethylethanolamine | ChEBI | | Choline cation | Kegg | | (2-Hydroxyethyl)trimethyl ammonium | HMDB | | (2-Hydroxyethyl)trimethylammonium | HMDB | | (beta-Hydroxyethyl)trimethylammonium | HMDB | | 2-Hydroxy-N,N,N-trimethyl-ethanaminium | HMDB | | 2-Hydroxy-N,N,N-trimethylethanaminium | HMDB | | Biocolina | HMDB | | Biocoline | HMDB | | Cholinum | HMDB | | Hepacholine | HMDB | | Hormocline | HMDB | | Lipotril | HMDB | | N,N,N-Trimethylethanolammonium | HMDB | | Neocolina | HMDB | | Paresan | HMDB | | Choline O-sulfate | HMDB | | Citrate, choline | HMDB | | Fagine | HMDB | | Hydroxide, choline | HMDB | | Vidine | HMDB | | Bursine | HMDB | | Chloride, choline | HMDB | | Choline chloride | HMDB | | Choline citrate | HMDB | | Bitartrate, choline | HMDB | | Choline bitartrate | HMDB | | Choline hydroxide | HMDB | | Choline O sulfate | HMDB | | O-Sulfate, choline | HMDB |
|
|---|
| Chemical Formula | C5H14NO |
|---|
| Average Molecular Weight | 104.17 |
|---|
| Monoisotopic Molecular Weight | 104.1075 |
|---|
| IUPAC Name | (2-hydroxyethyl)trimethylazanium |
|---|
| Traditional Name | choline |
|---|
| CAS Registry Number | 62-49-7 |
|---|
| SMILES | C[N+](C)(C)CCO |
|---|
| InChI Identifier | InChI=1S/C5H14NO/c1-6(2,3)4-5-7/h7H,4-5H2,1-3H3/q+1 |
|---|
| InChI Key | OEYIOHPDSNJKLS-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as cholines. These are organic compounds containing a N,N,N-trimethylethanolammonium cation. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Quaternary ammonium salts |
|---|
| Direct Parent | Cholines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Choline
- Tetraalkylammonium salt
- 1,2-aminoalcohol
- Alkanolamine
- Organic oxygen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Organic salt
- Primary alcohol
- Organooxygen compound
- Amine
- Alcohol
- Organic cation
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 302 - 303 °C (chloride salt) | Not Available | | Boiling Point | 113 °C | Wikipedia | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Choline, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-05fr-9100000000-69389426dd865af031f9 | Spectrum | | Predicted GC-MS | Choline, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-05g0-9300000000-ca1b0086d4882b00b395 | Spectrum | | Predicted GC-MS | Choline, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Choline, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0a4i-0900000000-ea6f9c1150759c5be6c7 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4i-1900000000-cadf16d18f093e9b19d6 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0bt9-9100000000-a2c328b7da5435b8e467 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0udi-0900000000-6a7b57dbdf4560721722 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-0udi-5900000000-87fdeef5275a74e78348 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0w29-5900000000-9b79cc5ff3b1ce17deb1 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-03di-9200000000-fe62d5a4fbf492493d60 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-0udi-3900000000-0b37e63e9c6b14a2e397 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0udi-0900000000-6a7b57dbdf4560721722 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-0udi-5900000000-87fdeef5275a74e78348 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , positive | splash10-0pb9-1900000000-391c1b7859a03e776123 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-2900000000-43b0f930ff04f7a48a59 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udr-9400000000-ab7cc75a8e0d3cc49449 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-9000000000-0b94edfd44e38b20b8d3 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-9100000000-a738404b52e2f1f6fec1 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-9000000000-c596e40769889223b3dd | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-9000000000-c6fab16d1790976560b5 | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Betaine Metabolism |  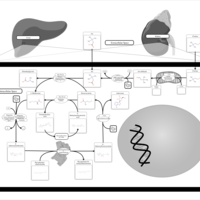  |  | | Methionine Metabolism | 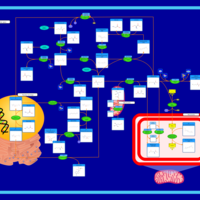 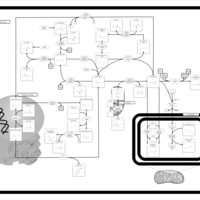  |  | | Phospholipid Biosynthesis |  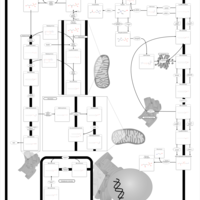 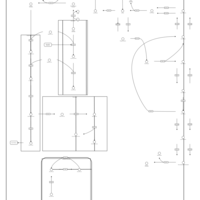 |  | | Cystathionine Beta-Synthase Deficiency | 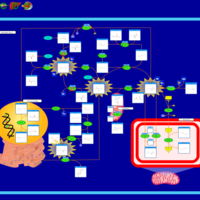 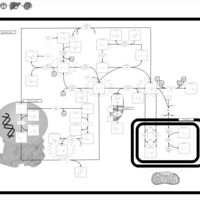  | Not Available | | Hypermethioninemia |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 1.418 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.853 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 1.128 mg/g dry wt | | details | | Quadra | Detected and Quantified | 1.00629 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.931 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 1.0750 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000097 |
|---|
| DrugBank ID | DB00122 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000710 |
|---|
| KNApSAcK ID | C00007298 |
|---|
| Chemspider ID | 299 |
|---|
| KEGG Compound ID | C00114 |
|---|
| BioCyc ID | CHOLINE |
|---|
| BiGG ID | 33910 |
|---|
| Wikipedia Link | Choline |
|---|
| METLIN ID | 56 |
|---|
| PubChem Compound | 305 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15354 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|


