| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-19 00:49:35 UTC |
|---|
| Updated at | 2020-12-07 19:07:37 UTC |
|---|
| CannabisDB ID | CDB000660 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Phenol |
|---|
| Description | Phenol, also known as hydroxybenzene or carbolic acid, belongs to the class of organic compounds known as hydroxy benzenoids. Phenol is a toxic, colourless crystalline solid with a sweet tarry odor that resembles a hospital smell. It is commonly used as an antiseptic and disinfectant. It is active against a wide range of microorganisms including some fungi and viruses but is only slowly effective against spores. It has been used to disinfect skin and to relieve itching. Phenol is also used in the preparation of cosmetics including sunscreens, hair dyes, and skin lightening preparations. It is also used in the production of drugs (it is the starting material in the industrial production of aspirin), weedkillers, and synthetic resins. Phenol can be found in areas with high levels of motor traffic, therefore, people living in crowded urban areas are frequently exposed to traffic-derived phenol vapor. The average (mean +/- SD) phenol concentration in urine among normal individuals living in urban areas is 7.4 +/- 2.2 mg/g of creatinine. Exposure of the skin to concentrated phenol solutions causes chemical burns which may be severe. In laboratories where it is used, it is usually recommended that polyethylene glycol solution is kept available for washing off splashes. In some bacteria phenol can be directly synthesized from tyrosine via the enzyme tyrosine phenol-lyase [EC:4.1.99.2]. It can be produced by Escherichia and Pseudomonas. Phenol has been identified as a uremic toxin according to the European Uremic Toxin Working Group (PMID: 22626821 ). Phenol also has been reported as a volatile compound detected in cannabis samples obtained through police seizures (PMID: 26657499 ). Phenol is also a constituent of cannabis plant and cannabis smoke. It is volatilized during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Acide carbolique | ChEBI | | Acide phenique | ChEBI | | Benzenol | ChEBI | | Carbolic acid | ChEBI | | Carbolsaeure | ChEBI | | Hydroxybenzene | ChEBI | | Karbolsaeure | ChEBI | | Oxybenzene | ChEBI | | Phenic acid | ChEBI | | Phenylic acid | ChEBI | | Phenylic alcohol | ChEBI | | PHOH | ChEBI | | Liquefied phenol | Kegg | | Phenol for disinfection | Kegg | | Phenol, liquefied | Kegg | | Paoscle | Kegg | | Carbolate | Generator | | Phenate | Generator | | Phenylate | Generator | | Anbesol | HMDB | | Benzophenol | HMDB | | Campho-phenique cold sore gel | HMDB | | Campho-phenique gel | HMDB | | Campho-phenique liquid | HMDB | | Carbolic acid liquid | HMDB | | Carbolic oil | HMDB | | Carbolicum acidum | HMDB | | Carbolsaure | HMDB | | Cepastat lozenges | HMDB | | Cuticura pain relieving ointment | HMDB | | Fenol | HMDB | | Fenolo | HMDB | | Fenosmolin | HMDB | | Fenosmoline | HMDB | | Hydroxy-benzene | HMDB | | IPH | HMDB | | IZAL | HMDB | | Liquid phenol | HMDB | | Liquified phenol | HMDB | | Monohydroxy benzene | HMDB | | Monohydroxybenzene | HMDB | | Monophenol | HMDB | | Phenic | HMDB | | Phenic alcohol | HMDB | | Phenol alcohol | HMDB | | Phenol homopolymer | HMDB | | Phenol liquid | HMDB | | Phenol molten | HMDB | | Phenol polymer-bound | HMDB | | Phenol solution | HMDB | | Phenol synthetic | HMDB | | Phenolated water | HMDB | | Phenolated water for disinfection | HMDB | | Phenole | HMDB | | Phenosmolin | HMDB | | Synthetic phenol | HMDB | | Tea polyphenol | HMDB | | Phenol, sodium salt | HMDB | | Phenolate sodium | HMDB | | Carbol | HMDB | | Phenolate, sodium | HMDB | | Sodium phenolate | HMDB |
|
|---|
| Chemical Formula | C6H6O |
|---|
| Average Molecular Weight | 94.11 |
|---|
| Monoisotopic Molecular Weight | 94.0419 |
|---|
| IUPAC Name | phenol |
|---|
| Traditional Name | phenol |
|---|
| CAS Registry Number | 108-95-2 |
|---|
| SMILES | OC1=CC=CC=C1 |
|---|
| InChI Identifier | InChI=1S/C6H6O/c7-6-4-2-1-3-5-6/h1-5,7H |
|---|
| InChI Key | ISWSIDIOOBJBQZ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 1-hydroxy-4-unsubstituted benzenoids. These are phenols that are unsubstituted at the 4-position. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Phenols |
|---|
| Sub Class | 1-hydroxy-4-unsubstituted benzenoids |
|---|
| Direct Parent | 1-hydroxy-4-unsubstituted benzenoids |
|---|
| Alternative Parents | |
|---|
| Substituents | - 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Monocyclic benzene moiety
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Source: Route of exposure: Biological location: |
|---|
| Role | Indirect biological role: Environmental role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 40.9 °C | Not Available | | Boiling Point | 181.7 °C | Wikipedia | | Water Solubility | 82.8 mg/mL at 25 °C | Not Available | | logP | 1.46 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-00kf-9000000000-40b376f4e58c23369a01 | 2014-09-20 | View Spectrum | | GC-MS | Phenol, 1 TMS, GC-MS Spectrum | splash10-0udi-3900000000-97dfa3be718a9ee7ace4 | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-00kf-9000000000-6fb456992902a13931f9 | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-78c83ab6ff1d3dfdbec6 | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-89b0c430b8924ee2afde | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-fc01d0ad6740cfd70e13 | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-006w-9000000000-c8b41f2899ca8cc9d4bc | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-0006-9000000000-db80d8b605e20595c679 | Spectrum | | GC-MS | Phenol, non-derivatized, GC-MS Spectrum | splash10-0udi-3900000000-97dfa3be718a9ee7ace4 | Spectrum | | Predicted GC-MS | Phenol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-c35c89484e2499c62a49 | Spectrum | | Predicted GC-MS | Phenol, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0g4i-9300000000-afd565a878ea36c74def | Spectrum | | Predicted GC-MS | Phenol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0006-9000000000-e67da0571423f0e6b4b8 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0006-9000000000-e0f57b1e970d0d46597e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0006-9000000000-e67da0571423f0e6b4b8 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI RMU-7M) , Positive | splash10-00kf-9000000000-7bfe3b897e928a8f3a2c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (JEOL JMS-01-SG-2) , Positive | splash10-0006-9000000000-78c83ab6ff1d3dfdbec6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI RMU-6L) , Positive | splash10-0006-9000000000-89b0c430b8924ee2afde | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (JEOL JMS-D-3000) , Positive | splash10-0006-9000000000-fc01d0ad6740cfd70e13 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-0006-9000000000-ca4fa5905cccbfeab33a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-e3e33defb000b450bb37 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9200000000-d63db79771e5931b258f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-448a4d79ff53d77a8b16 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-2c7201e803e029dd1aef | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-3fb3990dfbfbde2b8d57 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0gba-9000000000-d3da8db6579f32d02c6c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-a74494cda18ab9fb8055 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-a74494cda18ab9fb8055 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-03a61ff7da92cb08edf8 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9000000000-c65d4a4eaa8adde2ddb4 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-014j-9000000000-def7a8d52afb63a3cd67 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-f3c795ffc788635ddbf4 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-08485aaf085c13d5129a | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-08485aaf085c13d5129a | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-52b0f93832ef13f17bd2 | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Sulfate/Sulfite Metabolism | 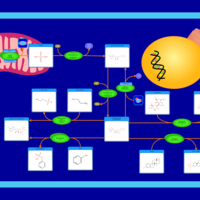 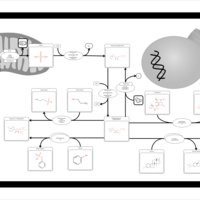  |  | | Sulfite oxidase deficiency | 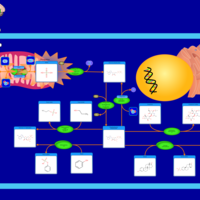 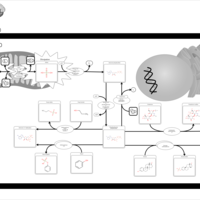  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000228 |
|---|
| DrugBank ID | DB03255 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000893 |
|---|
| KNApSAcK ID | C00002664 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C15584 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Phenol |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 996 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15882 |
|---|
| References |
|---|
| General References | - Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A: Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012 Jul;23(7):1258-70. doi: 10.1681/ASN.2011121175. Epub 2012 May 24. [PubMed:22626821 ]
- Rice S, Koziel JA: Characterizing the Smell of Marijuana by Odor Impact of Volatile Compounds: An Application of Simultaneous Chemical and Sensory Analysis. PLoS One. 2015 Dec 10;10(12):e0144160. doi: 10.1371/journal.pone.0144160. eCollection 2015. [PubMed:26657499 ]
|
|---|
