| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:25:49 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB000128 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Alanine |
|---|
| Description | L-Alanine or Alanine, abbreviated Ala or A, is a non-essential amino acid made in the body from either the conversion of the carbohydrate pyruvate or the breakdown of DNA and the dipeptides carnosine and anserine. Normal alanine metabolism, like that of other amino acids, is highly dependent upon enzymes that contain vitamin B6. Alanine is an important participant as well as a regulator of glucose metabolism. Alanine levels parallel blood sugar levels in both diabetes and hypoglycemia, and alanine reduces both severe hypoglycemia and the ketosis of diabetes. It is highly concentrated in muscle and is one of the most important amino acids released by muscle, functioning as a major energy source. Plasma alanine is often decreased when the BCAA (branched-chain amino acids) are deficient. This finding may relate to muscle metabolism. It is an important amino acid for lymphocyte reproduction and immunity. Alanine therapy has helped dissolve kidney stones in experimental animals. L-Alanine has been found to be associated with glucagon deficiency, which is an inborn error of metabolism. Alanine is highly concentrated in meat products and other high-protein foods like wheat germ and cottage cheese. L-alanine is also found in Cannabis plants (PMID: 6991645 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2-Aminopropanoic acid | ChEBI | | (S)-2-Aminopropanoic acid | ChEBI | | (S)-Alanine | ChEBI | | A | ChEBI | | Ala | ChEBI | | ALANINE | ChEBI | | L-2-Aminopropionic acid | ChEBI | | L-Alanin | ChEBI | | L-alpha-Alanine | ChEBI | | (2S)-2-Aminopropanoate | Generator | | (S)-2-Aminopropanoate | Generator | | L-2-Aminopropionate | Generator | | L-a-Alanine | Generator | | L-Α-alanine | Generator | | (S)-(+)-Alanine | HMDB | | (S)-2-Amino-propanoate | HMDB | | (S)-2-Amino-propanoic acid | HMDB | | 2-Aminopropanoate | HMDB | | 2-Aminopropanoic acid | HMDB | | 2-Aminopropionate | HMDB | | 2-Aminopropionic acid | HMDB | | 2-Ammoniopropanoate | HMDB | | 2-Ammoniopropanoic acid | HMDB | | a-Alanine | HMDB | | a-Aminopropionate | HMDB | | a-Aminopropionic acid | HMDB | | alpha-Alanine | HMDB | | alpha-Aminopropanoate | HMDB | | alpha-Aminopropanoic acid | HMDB | | alpha-Aminopropionate | HMDB | | alpha-Aminopropionic acid | HMDB | | L-(+)-Alanine | HMDB | | L-2-Aminopropanoate | HMDB | | L-2-Aminopropanoic acid | HMDB | | L-a-Aminopropionate | HMDB | | L-a-Aminopropionic acid | HMDB | | L-alpha-Aminopropionate | HMDB | | L-alpha-Aminopropionic acid | HMDB | | Abufène | HMDB | | Alanine doms-adrian brand | HMDB | | Alanine, L-isomer | HMDB | | Doms-adrian brand OF alanine | HMDB | | Doms adrian brand OF alanine | HMDB | | L-Isomer alanine | HMDB | | Alanine, L isomer | HMDB | | L Alanine | HMDB |
|
|---|
| Chemical Formula | C3H7NO2 |
|---|
| Average Molecular Weight | 89.09 |
|---|
| Monoisotopic Molecular Weight | 89.0477 |
|---|
| IUPAC Name | (2S)-2-aminopropanoic acid |
|---|
| Traditional Name | L-alanine |
|---|
| CAS Registry Number | 115967-49-2 |
|---|
| SMILES | C[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m0/s1 |
|---|
| InChI Key | QNAYBMKLOCPYGJ-REOHCLBHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as alanine and derivatives. Alanine and derivatives are compounds containing alanine or a derivative thereof resulting from reaction of alanine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Alanine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Alanine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Hydrocarbon derivative
- Organic oxygen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Amine
- Organopnictogen compound
- Organic oxide
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 300 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 204 mg/mL | Human Metabolome Project | | logP | -2.85 | SANGSTER (1994) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Alanine, 1 TMS, GC-MS Spectrum | splash10-014i-0900000000-c7f6dbace291e8305a4e | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-014i-0900000000-381ddf4d9ea77be0b8a5 | Spectrum | | GC-MS | L-Alanine, 3 TMS, GC-MS Spectrum | splash10-01b9-6900000000-6a7c1bb2915e5dd0791f | Spectrum | | GC-MS | L-Alanine, 2 TMS, GC-MS Spectrum | splash10-014i-1900000000-84b389f82562c29a8148 | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-00kf-9000000000-b2f7507be509a85d821b | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-014i-0900000000-941672891ab94cf5015d | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-0f79-0910000000-47bd3e3aa274a653a64e | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-014i-0900000000-c7f6dbace291e8305a4e | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-014i-0900000000-381ddf4d9ea77be0b8a5 | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-0a4i-1940000000-5def9f7c902aaf1ef607 | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-01b9-6900000000-6a7c1bb2915e5dd0791f | Spectrum | | GC-MS | L-Alanine, non-derivatized, GC-MS Spectrum | splash10-014i-1900000000-84b389f82562c29a8148 | Spectrum | | Predicted GC-MS | L-Alanine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-d31f7a2ed8284a740b59 | Spectrum | | Predicted GC-MS | L-Alanine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9100000000-ab365202f52df8e6d401 | Spectrum | | Predicted GC-MS | L-Alanine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Alanine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Alanine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Alanine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-000i-9000000000-55d0139f513946f76461 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-000i-9000000000-5b0dff13a98daf782205 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-000i-9000000000-37db595fcf7364600bc9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-55d0139f513946f76461 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-5b0dff13a98daf782205 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-000i-9000000000-37db595fcf7364600bc9 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-000i-9000000000-9ca90486c916f938450b | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-000i-9000000000-e9e9900761fdd599ee3f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-17dcfdd88dc79713c4b7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-000i-9000000000-8cc23f8c1455fdf68df5 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000i-9000000000-7b5177095a55ce689565 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 33V, Negative | splash10-0uxr-9400000000-b26be1c8f3385f90fcc3 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 33V, Negative | splash10-02ti-9000000000-41eecbbaba19b8b0b536 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0006-9000000000-96b54b269c91ab21be08 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0006-9000000000-a8008305399aa1097e1a | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0006-9000000000-7253c912562200edc231 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI RMU-6M) , Positive | splash10-00kf-9000000000-72694f3a1a5de3b49790 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0006-9000000000-5129e160acf979ac549e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0006-9000000000-11c20eba8c5ad41d806c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0006-9000000000-df7c7d6a6ae2d6bccefe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0006-9000000000-32477244247613182214 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-000f-9002000000-4873ce0cdcac54d68186 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-0006-9000000000-8193842b36c819ca3ec6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0006-9000000000-fefc31c264a2f0fcf6d0 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0006-9000000000-ba42bcd60aa9182a780e | 2017-09-14 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Urea Cycle | 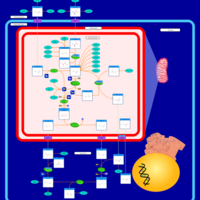 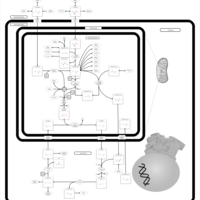 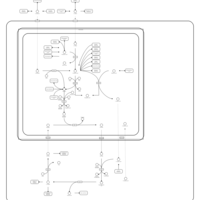 |  | | Glucose-Alanine Cycle | 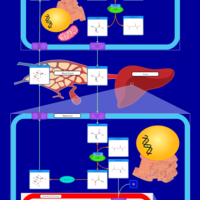 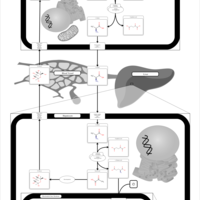 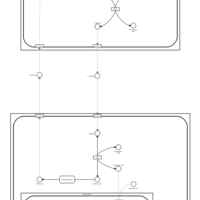 | Not Available | | Selenoamino Acid Metabolism |  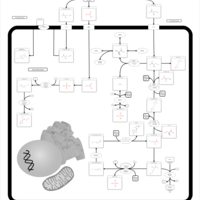  |  | | Alanine Metabolism | 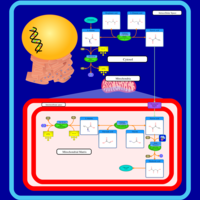 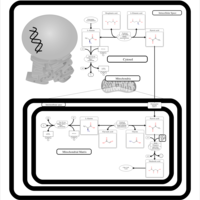  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Large neutral amino acids transporter small subunit 2 | SLC7A8 | 14q11.2 | Q9UHI5 | details | | Sodium-coupled neutral amino acid transporter 3 | SLC38A3 | 3p21.3 | Q99624 | details | | Neutral amino acid transporter A | SLC1A4 | 2p15-p13 | P43007 | details | | Proton-coupled amino acid transporter 1 | SLC36A1 | 5q33.1 | Q7Z2H8 | details | | Monocarboxylate transporter 10 | SLC16A10 | 6q21-q22 | Q8TF71 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.284 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.337 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.462 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.315 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.452 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.432 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000161 |
|---|
| DrugBank ID | DB00160 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000556 |
|---|
| KNApSAcK ID | C00001332 |
|---|
| Chemspider ID | 5735 |
|---|
| KEGG Compound ID | C00041 |
|---|
| BioCyc ID | L-ALPHA-ALANINE |
|---|
| BiGG ID | 33629 |
|---|
| Wikipedia Link | Alanine |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 5950 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16977 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|


