| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:45:37 UTC |
|---|
| Updated at | 2021-01-04 18:59:17 UTC |
|---|
| CannabisDB ID | CDB004873 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Pyrocatechol |
|---|
| Description | Pyrocatechol, or o-Benzenediol, often known as catechol or benzene-1,2-diol, is a benzenediol. It belongs to the class of organic compounds known as catechols. Catechols are compounds containing a 1,2-benzenediol moiety. Pyrocatechol is one of three known isomers of benzenediol including o-Benzenediol (Catechol), m-benzenediol (Resorcinol) and p-benzenediol (Hydroquinone). Pyrocatechol exists as white crystals and is very soluble in water. It is also soluble in pyridine and other organic solvents such as chloroform, benzene, CCl4, ether, and ethyl acetate. Pyrocatechol was first prepared in 1839 by H. Reinsch by distilling catechin (the juice of Mimosa catechu). This colourless compound occurs naturally, but about 20000 tons are manufactured each year, mainly as precursors to pesticides, flavors, and fragrances. Small amounts of catechol occur naturally in fruits and vegetables, along with the enzyme polyphenol oxidase. Upon mixing the enzyme with the substrate and exposure to oxygen (as when a potato or apple is cut), the colorless catechol oxidizes to reddish-brown benzoquinone derivatives. The enzyme is inactivated by adding an acid, such as lemon juice, or by refrigeration. Excluding oxygen also prevents the browning reaction. Catechol is produced industrially by the hydroxylation of phenol using hydrogen peroxide ( Ref:DOI ). Catechol is used in medicine as an expectorant. The dimethyl ether or veratrol is also used in medicine. Many other pyrocatechin derivatives have been suggested for therapeutic application. Its sulfonic acid is often present in the urine of many mammals (PMID: 18499097 ). Pyrocatechol has also been found to be a microbial metabolite in Escherichia, Mycobacterium and Pseudomonas (PMID: 19300498 ; PMID: 25281236 ). Catechol is one of several phenolic compounds found in cannabis smoke and is formed during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,2-Benzenediol | ChEBI | | 1,2-Dihydroxybenzene | ChEBI | | 2-Hydroxyphenol | ChEBI | | alpha-Hydroxyphenol | ChEBI | | Brenzcatechin | ChEBI | | O-Benzenediol | ChEBI | | O-Hydroxyphenol | ChEBI | | Pyrocatechin | ChEBI | | a-Hydroxyphenol | Generator | | Α-hydroxyphenol | Generator | | Catechol | HMDB | | Durafur developer C | HMDB | | Fouramine PCH | HMDB | | Fourrine 68 | HMDB | | O-Dihydroxybenzene | HMDB | | O-Dioxybenzene | HMDB | | O-Hydroquinone | HMDB | | O-Phenylenediol | HMDB | | Oxyphenate | HMDB | | Oxyphenic acid | HMDB | | Pelagol grey C | HMDB | | Phthalhydroquinone | HMDB | | Phthalic alcohol | HMDB | | Pyrocatechine | HMDB | | 1,3-Dihydroxybenzene | HMDB | | Catechol dipotassium salt | HMDB | | Catechol, 14C-labeled CPD | HMDB | | Catechol sodium salt | HMDB | | Benzene-1,2-diol | PhytoBank | | Pyrocatechol | PhytoBank | | 1,2-Hydroxybenzene | PhytoBank |
|
|---|
| Chemical Formula | C6H6O2 |
|---|
| Average Molecular Weight | 110.11 |
|---|
| Monoisotopic Molecular Weight | 110.0368 |
|---|
| IUPAC Name | benzene-1,2-diol |
|---|
| Traditional Name | catechol |
|---|
| CAS Registry Number | 120-80-9 |
|---|
| SMILES | OC1=CC=CC=C1O |
|---|
| InChI Identifier | InChI=1S/C6H6O2/c7-5-3-1-2-4-6(5)8/h1-4,7-8H |
|---|
| InChI Key | YCIMNLLNPGFGHC-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as catechols. Catechols are compounds containing a 1,2-benzenediol moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Phenols |
|---|
| Sub Class | Benzenediols |
|---|
| Direct Parent | Catechols |
|---|
| Alternative Parents | |
|---|
| Substituents | - Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- 1-hydroxy-2-unsubstituted benzenoid
- Monocyclic benzene moiety
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Environmental role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 105 °C | Not Available | | Boiling Point | 245.5 °C | Wikipedia | | Water Solubility | 461 mg/mL | Not Available | | logP | 0.88 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-03di-9600000000-b8e03f4f3ea89044828e | 2014-09-20 | View Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-0udr-1950000000-16187bb35dcb40c26e78 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-03di-8900000000-4e15f35dca47661de590 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-03di-9600000000-032a40483dec93738075 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-03di-7900000000-83f892852c355a3863e9 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-03di-9400000000-90885264baa17f65d954 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-0udr-1950000000-16187bb35dcb40c26e78 | Spectrum | | GC-MS | Pyrocatechol, non-derivatized, GC-MS Spectrum | splash10-0udr-1930000000-24d2e0a8e36245d9e187 | Spectrum | | Predicted GC-MS | Pyrocatechol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03di-7900000000-8b112b8af75eb2d08676 | Spectrum | | Predicted GC-MS | Pyrocatechol, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fl9-9730000000-a35befff1c602124f29e | Spectrum | | Predicted GC-MS | Pyrocatechol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Pyrocatechol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Pyrocatechol, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Pyrocatechol, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Pyrocatechol, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-01ox-9400000000-d59dce8c5e56b026f8b2 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-014i-9000000000-632cabc9b371835019c1 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-02t9-9200000000-ac902cb99981017de3b5 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (VARIAN MAT-44) , Positive | splash10-03di-8900000000-95af3d2738de98d27f26 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (Unknown) , Positive | splash10-03di-9600000000-032a40483dec93738075 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80) , Positive | splash10-03di-7900000000-f5cb1c53768e05ca1530 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0a4i-0900000000-c94dab4d218dbb3bb108 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Negative | splash10-0a4i-1900000000-edd8ba1e77bbb2f76304 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-0900000000-c94dab4d218dbb3bb108 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-1900000000-edd8ba1e77bbb2f76304 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-0a4i-0900000000-12053747e62e910151ad | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-72e952ea8e487994be54 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-19b1bae28f3dfde3323a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0006-9000000000-33b2a7e7a951547dfafa | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-6e731d4eaba18fcad18f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-1900000000-a7c1a830ea96e82252bf | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-3772b2cca96bf4a1b05f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-9531f3e0c85e67d6c6ed | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-1900000000-5886b926a1814092c4b1 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-66523f3122b954e6400f | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-1900000000-5fd776e479836f7464af | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0udi-9000000000-2c46a1375dbb634ef735 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-986c93875cb12d90fa90 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-a301685abb4194689ca3 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9500000000-2017b42835ace86f16ee | 2015-05-27 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Tyrosine Metabolism | 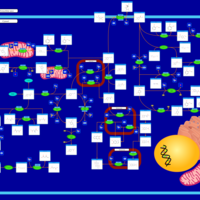 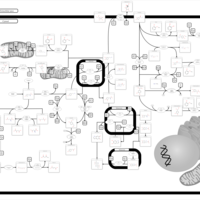 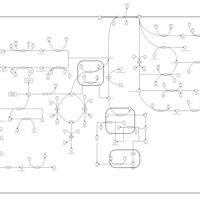 |  | | Alkaptonuria | 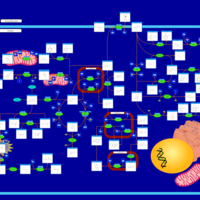 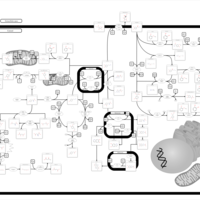 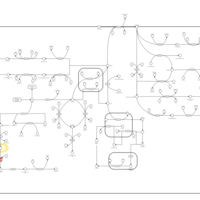 | Not Available | | Hawkinsinuria | 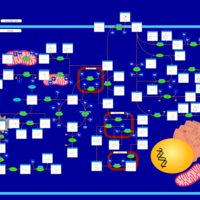  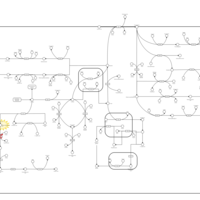 | Not Available | | Tyrosinemia Type I | 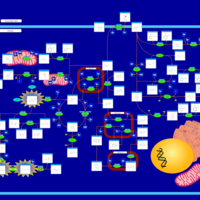 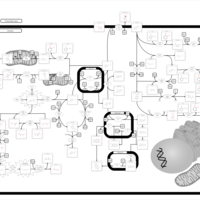 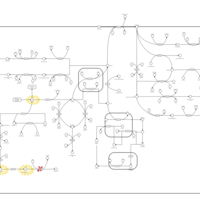 | Not Available | | Tyrosinemia, transient, of the newborn | 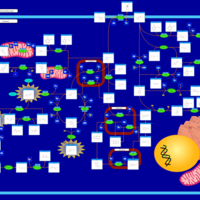 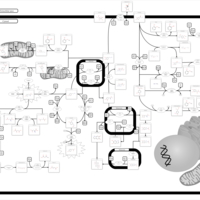 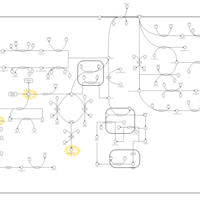 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Aldo-keto reductase family 1 member C4 | AKR1C4 | 10p15.1 | P17516 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000957 |
|---|
| DrugBank ID | DB02232 |
|---|
| Phenol Explorer Compound ID | 654 |
|---|
| FoodDB ID | FDB001512 |
|---|
| KNApSAcK ID | C00002644 |
|---|
| Chemspider ID | 13837760 |
|---|
| KEGG Compound ID | C15571 |
|---|
| BioCyc ID | CATECHOL |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Catechol |
|---|
| METLIN ID | 282 |
|---|
| PubChem Compound | 289 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 18135 |
|---|
| References |
|---|
| General References | - Zheng LT, Ryu GM, Kwon BM, Lee WH, Suk K: Anti-inflammatory effects of catechols in lipopolysaccharide-stimulated microglia cells: inhibition of microglial neurotoxicity. Eur J Pharmacol. 2008 Jun 24;588(1):106-13. doi: 10.1016/j.ejphar.2008.04.035. Epub 2008 Apr 18. [PubMed:18499097 ]
- Yam KC, D'Angelo I, Kalscheuer R, Zhu H, Wang JX, Snieckus V, Ly LH, Converse PJ, Jacobs WR Jr, Strynadka N, Eltis LD: Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 2009 Mar;5(3):e1000344. doi: 10.1371/journal.ppat.1000344. Epub 2009 Mar 20. [PubMed:19300498 ]
- Balderas-Hernandez VE, Trevino-Quintanilla LG, Hernandez-Chavez G, Martinez A, Bolivar F, Gosset G: Catechol biosynthesis from glucose in Escherichia coli anthranilate-overproducer strains by heterologous expression of anthranilate 1,2-dioxygenase from Pseudomonas aeruginosa PAO1. Microb Cell Fact. 2014 Oct 4;13:136. doi: 10.1186/s12934-014-0136-x. [PubMed:25281236 ]
|
|---|
