| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:43:27 UTC |
|---|
| Updated at | 2020-12-07 19:11:06 UTC |
|---|
| CannabisDB ID | CDB004852 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Dodecanoic acid |
|---|
| Description | Dodecanoic acid also known as Lauric acid, belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. Lauric acid is a weakly acidic compound. It is a white, powdery solid with a faint odour of mild fatty coconut bay oil or soap. Dodecanoic acid is the main fatty acid in coconut oil (49%) and in palm kernel oil (47-50%), and is found in lesser amounts in wild nutmeg, human breast milk, cow’s milk, goat milk, watermelon seeds, plum and macadamia nut ( doi:10.1351/pac200173040685). Dodecanoic acid is one of the saturated fatty acids found in the oil of cannabis seeds ( Ref:DOI ). Dodecanoic acid, although slightly irritating to mucous membranes, has an extremely low toxicity, is inexpensive, has antimicrobial properties and so is used in many soaps and shampoos. It is reacted with sodium hydroxide to generate sodium laurate, which is soap. Dodecanoic acid has been characterized as having "a more favorable effect on total HDL cholesterol than any other fatty acid either saturated or unsaturated" (PMID: 12716665 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Undecanecarboxylic acid | ChEBI | | ABL | ChEBI | | C12 Fatty acid | ChEBI | | C12:0 | ChEBI | | CH3-[CH2]10-COOH | ChEBI | | Coconut oil fatty acids | ChEBI | | DAO | ChEBI | | Dodecoic acid | ChEBI | | Dodecylcarboxylate | ChEBI | | Dodecylic acid | ChEBI | | Duodecyclic acid | ChEBI | | Duodecylic acid | ChEBI | | Lauric acid | ChEBI | | Laurinsaeure | ChEBI | | Laurostearic acid | ChEBI | | N-Dodecanoic acid | ChEBI | | Undecane-1-carboxylic acid | ChEBI | | Vulvic acid | ChEBI | | Dodecanoate | Kegg | | 1-Undecanecarboxylate | Generator | | Dodecoate | Generator | | Dodecylcarboxylic acid | Generator | | Dodecylate | Generator | | Duodecyclate | Generator | | Duodecylate | Generator | | Laate | Generator | | Laic acid | Generator | | Laurostearate | Generator | | N-Dodecanoate | Generator | | Undecane-1-carboxylate | Generator | | Vulvate | Generator | | Aliphat no. 4 | HMDB | | Edenor C 1298-100 | HMDB | | Emery 651 | HMDB | | Hystrene 9512 | HMDB | | Kortacid 1299 | HMDB | | Laurate | HMDB | | Lunac L 70 | HMDB | | Lunac L 98 | HMDB | | Neo-fat 12 | HMDB | | Neo-fat 12-43 | HMDB | | Nissan naa 122 | HMDB | | Philacid 1200 | HMDB | | Prifac 2920 | HMDB | | Univol u 314 | HMDB | | 1-Dodecanoic acid | HMDB | | FA(12:0) | HMDB | | Dodecanoic acid | PhytoBank |
|
|---|
| Chemical Formula | C12H24O2 |
|---|
| Average Molecular Weight | 200.32 |
|---|
| Monoisotopic Molecular Weight | 200.1776 |
|---|
| IUPAC Name | dodecanoic acid |
|---|
| Traditional Name | lauric acid |
|---|
| CAS Registry Number | 143-07-7 |
|---|
| SMILES | CCCCCCCCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C12H24O2/c1-2-3-4-5-6-7-8-9-10-11-12(13)14/h2-11H2,1H3,(H,13,14) |
|---|
| InChI Key | POULHZVOKOAJMA-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Medium-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 44 - 46 °C | Not Available | | Boiling Point | 282.5 °C at 512 mmHg | Wikipedia | | Water Solubility | 0.0048 mg/mL | Not Available | | logP | 4.60 | SANGSTER (1993) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-074l-9100000000-911a01d38925d0de481f | 2014-09-20 | View Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-a12b321a54a44ae28972 | Spectrum | | GC-MS | Dodecanoic acid, 1 TMS, GC-MS Spectrum | splash10-014i-2910000000-d52410168e784872a5a6 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-06xx-9100000000-1ef5cd411f38c53a0a92 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-0706-9100000000-e66a9147257053b93702 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-0706-9100000000-2dd666a9a0355b1ca144 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-074i-9300000000-405ec308fe8c935c62a2 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-067i-0930000000-6a80d41e346f4b10104c | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-a12b321a54a44ae28972 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-014i-2910000000-d52410168e784872a5a6 | Spectrum | | GC-MS | Dodecanoic acid, non-derivatized, GC-MS Spectrum | splash10-0159-1910000000-8246864bf316bce609a2 | Spectrum | | Predicted GC-MS | Dodecanoic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9500000000-e05832f94ff0361363a2 | Spectrum | | Predicted GC-MS | Dodecanoic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-05g0-9310000000-038e232cccb44582ec9c | Spectrum | | Predicted GC-MS | Dodecanoic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Dodecanoic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-2290000000-861c4b7b2da35b82ef7f | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4l-9100000000-966cf4c6621fcec41368 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-054o-9000000000-d43dd32656ad22e95fdd | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-06xx-9100000000-a71409efaf5fdd6ab0e1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (JEOL JMS-AX-505-H) , Positive | splash10-0706-9100000000-eb09296a8c4c7c381b38 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-f22d8298d64e6f382279 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-0900000000-4bb88d3b1d2b7ac88152 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0002-0900000000-0c79a7257b53685108c6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0002-9600000000-25d268515337ef276c60 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-009b-9000000000-c00963f9b91f4a82e1ef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0900000000-f22d8298d64e6f382279 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0900000000-4bb88d3b1d2b7ac88152 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0002-0900000000-0c79a7257b53685108c6 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0920000000-63cd88b9477cd281d342 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a5c-4910000000-94738a1f436dcf1abced | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9100000000-07ca8cffe5ba2f9f920d | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-0920000000-63cd88b9477cd281d342 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a5c-4910000000-94738a1f436dcf1abced | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9100000000-07ca8cffe5ba2f9f920d | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-1655e63166b4d9dd4f59 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0532-1900000000-7c55a9ca2b49d4920029 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9400000000-ed8f6bcf96c68ac84738 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-1655e63166b4d9dd4f59 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0532-1900000000-7c55a9ca2b49d4920029 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9400000000-ed8f6bcf96c68ac84738 | 2015-05-27 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 90 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, CDCl3, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, CD3OD, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Beta Oxidation of Very Long Chain Fatty Acids | 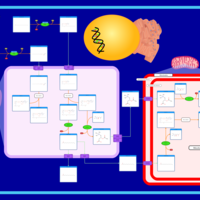 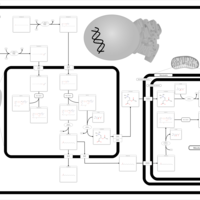 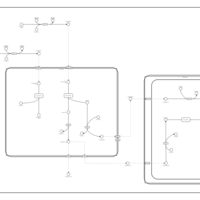 |  | | Fatty Acid Biosynthesis | 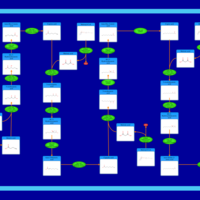 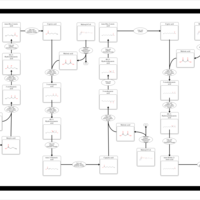 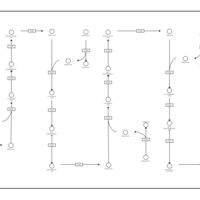 | Not Available | | Mitochondrial Beta-Oxidation of Medium Chain Saturated Fatty Acids | 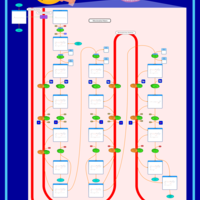 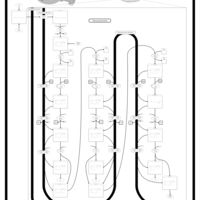 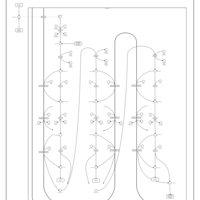 | Not Available | | Adrenoleukodystrophy, X-linked | 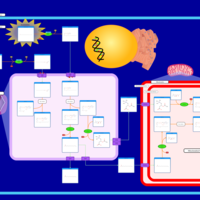 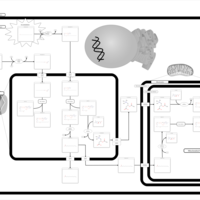 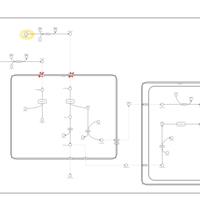 | Not Available | | Carnitine-acylcarnitine translocase deficiency |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Mitochondrial carnitine/acylcarnitine carrier protein | SLC25A20 | 3p21.31 | O43772 | details | | Glycolipid transfer protein | GLTP | 12q24.11 | Q9NZD2 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000638 |
|---|
| DrugBank ID | DB03017 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB003010 |
|---|
| KNApSAcK ID | C00001221 |
|---|
| Chemspider ID | 3756 |
|---|
| KEGG Compound ID | C02679 |
|---|
| BioCyc ID | DODECANOATE |
|---|
| BiGG ID | 40351 |
|---|
| Wikipedia Link | Lauric_acid |
|---|
| METLIN ID | 5611 |
|---|
| PubChem Compound | 3893 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30805 |
|---|
| References |
|---|
| General References | - Mensink RP, Zock PL, Kester AD, Katan MB: Effects of dietary fatty acids and carbohydrates on the ratio of serum total to HDL cholesterol and on serum lipids and apolipoproteins: a meta-analysis of 60 controlled trials. Am J Clin Nutr. 2003 May;77(5):1146-55. doi: 10.1093/ajcn/77.5.1146. [PubMed:12716665 ]
|
|---|
