| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:41:53 UTC |
|---|
| Updated at | 2020-11-18 16:38:49 UTC |
|---|
| CannabisDB ID | CDB004837 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Uridine diphosphate glucose |
|---|
| Description | Uridine diphosphate glucose, also known as udpglucose or UDP-alpha-D-glucose, belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. Uridine diphosphate glucose is an extremely weak basic (essentially neutral) compound (based on its pKa). Uridine diphosphate glucose exists in all living species, ranging from bacteria to humans. Within humans, uridine diphosphate glucose participates in a number of enzymatic reactions. In particular, galactose 1-phosphate and uridine diphosphate glucose can be biosynthesized from uridine diphosphategalactose and glucose 1-phosphate; which is catalyzed by the enzyme galactose-1-phosphate uridylyltransferase. In addition, uridine diphosphate glucose can be biosynthesized from uridine diphosphategalactose through its interaction with the enzyme UDP-glucose 4-epimerase. In humans, uridine diphosphate glucose is involved in galactose metabolism. Outside of the human body, Uridine diphosphate glucose has been detected, but not quantified in, several different foods, such as tea, ostrich ferns, chickpea, cinnamons, and sourdoughs. This could make uridine diphosphate glucose a potential biomarker for the consumption of these foods. Uridine diphosphate glucose is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| GLUCOSE-uridine-C1,5'-diphosphATE | ChEBI | | UDP-D-Glucose | ChEBI | | UDPglucose | ChEBI | | URIDINE-5'-diphosphATE-glucose | ChEBI | | UDP-alpha-D-Glucose | Kegg | | GLUCOSE-uridine-C1,5'-diphosphoric acid | Generator | | URIDINE-5'-diphosphoric acid-glucose | Generator | | UDP-a-D-Glucose | Generator | | UDP-Α-D-glucose | Generator | | Uridine diphosphoric acid glucose | Generator | | Diphosphate glucose, uridine | HMDB | | Diphosphoglucose, uridine | HMDB | | Glucose, UDP | HMDB | | Glucose, uridine diphosphate | HMDB | | UDP Glucose | HMDB | | UDPG | HMDB | | Uridine diphosphoglucose | HMDB | | UDP-GLC | HMDB | | UDP-Glucose | HMDB | | Uridine 5'-(alpha-D-glucopyranosyl pyrophosphate) | HMDB | | Uridine 5'-(α-D-glucopyranosyl pyrophosphate) | HMDB | | Uridine 5'-diphosphate glucose | HMDB | | Uridine 5'-diphospho-alpha-D-glucose | HMDB | | Uridine 5'-diphospho-α-D-glucose | HMDB | | Uridine 5'-diphosphoglucose | HMDB | | Uridine 5’-(α-D-glucopyranosyl pyrophosphate) | HMDB | | Uridine 5’-diphosphate glucose | HMDB | | Uridine 5’-diphospho-α-D-glucose | HMDB | | Uridine 5’-diphosphoglucose | HMDB | | Uridine diphospho-D-glucose | HMDB | | Uridine pyrophosphate-glucose | HMDB | | Uridine diphosphate glucose | ChEBI |
|
|---|
| Chemical Formula | C15H24N2O17P2 |
|---|
| Average Molecular Weight | 566.3 |
|---|
| Monoisotopic Molecular Weight | 566.055 |
|---|
| IUPAC Name | [({[(2R,3S,4R,5R)-5-(2,4-dioxo-1,2,3,4-tetrahydropyrimidin-1-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy]({[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy})phosphinic acid |
|---|
| Traditional Name | udp-α-D-glucose |
|---|
| CAS Registry Number | 133-89-1 |
|---|
| SMILES | OC[C@H]1O[C@H](OP(O)(=O)OP(O)(=O)OC[C@H]2O[C@H]([C@H](O)[C@@H]2O)N2C=CC(=O)NC2=O)[C@H](O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C15H24N2O17P2/c18-3-5-8(20)10(22)12(24)14(32-5)33-36(28,29)34-35(26,27)30-4-6-9(21)11(23)13(31-6)17-2-1-7(19)16-15(17)25/h1-2,5-6,8-14,18,20-24H,3-4H2,(H,26,27)(H,28,29)(H,16,19,25)/t5-,6-,8-,9-,10+,11-,12-,13-,14-/m1/s1 |
|---|
| InChI Key | HSCJRCZFDFQWRP-JZMIEXBBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pyrimidine nucleotide sugars. These are pyrimidine nucleotides bound to a saccharide derivative through the terminal phosphate group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | Pyrimidine nucleotides |
|---|
| Sub Class | Pyrimidine nucleotide sugars |
|---|
| Direct Parent | Pyrimidine nucleotide sugars |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrimidine nucleotide sugar
- Pyrimidine ribonucleoside diphosphate
- Pentose phosphate
- Pentose-5-phosphate
- Glycosyl compound
- N-glycosyl compound
- Monosaccharide phosphate
- Organic pyrophosphate
- Pyrimidone
- Monoalkyl phosphate
- Hydropyrimidine
- Monosaccharide
- Organic phosphoric acid derivative
- Oxane
- Phosphoric acid ester
- Pyrimidine
- Alkyl phosphate
- Heteroaromatic compound
- Vinylogous amide
- Tetrahydrofuran
- Urea
- Secondary alcohol
- Lactam
- Polyol
- Oxacycle
- Organoheterocyclic compound
- Azacycle
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Primary alcohol
- Alcohol
- Organonitrogen compound
- Organooxygen compound
- Organic oxygen compound
- Organopnictogen compound
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Uridine diphosphate glucose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-054k-3933570000-89f658b2d26ff158d84c | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fk9-4942706000-3a83736296429f83adee | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, "Uridine diphosphate glucose,1TMS,#1" TMS, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_1_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_11, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_12, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_13, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Uridine diphosphate glucose, TMS_2_14, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-014i-0000090000-1692ab5f5dad1cf7e436 | 2019-05-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-015a-0439510000-69e7cd4c2c365a24706b | 2019-05-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0109-5753970000-03f34041208ba93ef87a | 2019-05-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00fr-9257000000-73b85a465d99ce92467d | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-00ba-9333000000-b57dc1340555f1a997b4 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-00fr-9257000000-ba1e6c754b8f04d5b659 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0002-9000000000-9bd63595c19dbe18d143 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-00di-0009000000-4b94aa10fd96d2d9c505 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-00di-0009000000-996cd4acdaa34c877460 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0901110000-4b81825405869909992e | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-3911000000-6fd09254750e77963110 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-5900000000-03a2a98eb0aa84737530 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-4901260000-8396a090c14de7f2d708 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01ox-8907020000-94410556a0288cb8167b | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-06tf-5901000000-10132c85ad33464d4eef | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-0000960000-6404d8d944e1ae6f30e2 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-07e1-2322960000-6af539ce21752f9023ba | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-08fr-9732000000-b0c81adc41b1d799e1e8 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-0000090000-8cab8f2bc0fb9e9e8ba9 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-057j-9203870000-d60681ff91f741bdb867 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-3519000000-67ca2f9355ef7f94a47d | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Starch and Sucrose Metabolism | 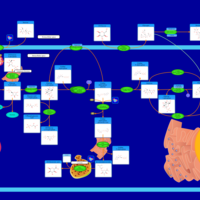 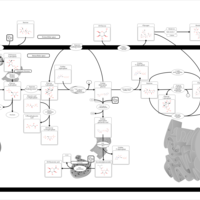 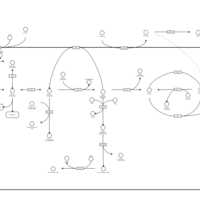 |  | | Lactose Synthesis | 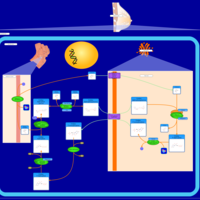 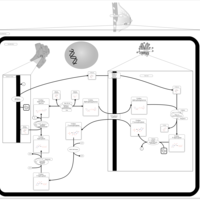 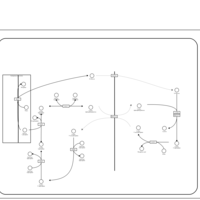 | Not Available | | Galactose Metabolism |  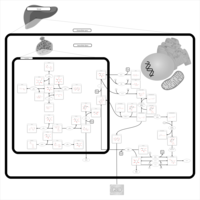 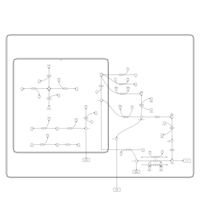 |  | | Nucleotide Sugars Metabolism | 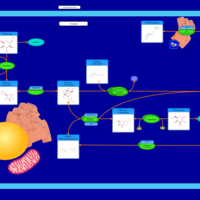 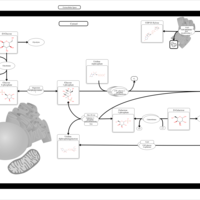 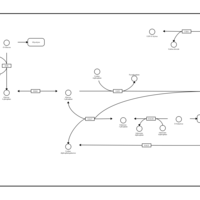 |  | | Galactosemia II (GALK) | 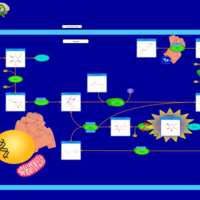 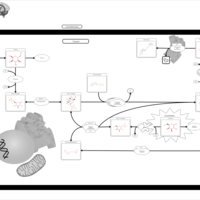 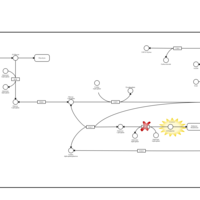 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | ENPP1 | 6q22-q23 | P22413 | details | | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | ENPP3 | 6q22 | O14638 | details | | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 3 | PLOD3 | 7q22 | O60568 | details | | UTP--glucose-1-phosphate uridylyltransferase | UGP2 | 2p14-p13 | Q16851 | details | | Galactose-1-phosphate uridylyltransferase | GALT | 9p13 | P07902 | details | | Glycogenin-1 | GYG1 | 3q24-q25.1 | P46976 | details | | Uridine diphosphate glucose pyrophosphatase | NUDT14 | 14q32.33 | O95848 | details |
|
|---|
| Receptors | |
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 | ENPP1 | 6q22-q23 | P22413 | details | | Ectonucleotide pyrophosphatase/phosphodiesterase family member 3 | ENPP3 | 6q22 | O14638 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000286 |
|---|
| DrugBank ID | DB01861 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB005660 |
|---|
| KNApSAcK ID | C00001514 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00029 |
|---|
| BioCyc ID | CPD-12575 |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Uridine_diphosphate_glucose |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 8629 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 46229 |
|---|
| References |
|---|
| General References | Not Available |
|---|


