| Identification |
|---|
| HMDB Protein ID
| CDBP00114 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ectonucleotide pyrophosphatase/phosphodiesterase family member 1 |
| Description
| Not Available |
| Synonyms
|
- Alkaline phosphodiesterase I
- E-NPP 1
- Membrane component chromosome 6 surface marker 1
- NPPase
- Nucleotide pyrophosphatase
- Phosphodiesterase I/nucleotide pyrophosphatase 1
- Plasma-cell membrane glycoprotein PC-1
|
| Gene Name
| ENPP1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Involved primarily in ATP hydrolysis at the plasma membrane. Plays a role in regulating pyrophosphate levels, and functions in bone mineralization and soft tissue calcification. In vitro, has a broad specificity, hydrolyzing other nucleoside 5' triphosphates such as GTP, CTP, TTP and UTP to their corresponding monophosphates with release of pyrophosphate and diadenosine polyphosphates, and also 3',5'-cAMP to AMP. May also be involved in the regulation of the availability of nucleotide sugars in the endoplasmic reticulum and Golgi, and the regulation of purinergic signaling. Appears to modulate insulin sensitivity.
|
| GO Classification
|
| Biological Process |
| negative regulation of fat cell differentiation |
| negative regulation of glucose import |
| negative regulation of glycogen biosynthetic process |
| negative regulation of insulin receptor signaling pathway |
| negative regulation of protein autophosphorylation |
| riboflavin metabolic process |
| sequestering of triglyceride |
| cellular response to insulin stimulus |
| generation of precursor metabolites and energy |
| regulation of bone mineralization |
| 3'-phosphoadenosine 5'-phosphosulfate metabolic process |
| nucleoside triphosphate catabolic process |
| immune response |
| biomineral tissue development |
| negative regulation of cell growth |
| bone remodeling |
| negative regulation of ossification |
| cellular phosphate ion homeostasis |
| inorganic diphosphate transport |
| Cellular Component |
| extracellular space |
| integral to membrane |
| cell surface |
| basolateral plasma membrane |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| catalytic activity |
| hydrolase activity |
| pattern binding |
| polysaccharide binding |
| receptor activity |
| transmembrane receptor activity |
| scavenger receptor activity |
| molecular transducer activity |
| signal transducer activity |
| nucleic acid binding |
| Molecular Function |
| nucleic acid binding |
| nucleoside-triphosphate diphosphatase activity |
| metal ion binding |
| insulin receptor binding |
| NADH pyrophosphatase activity |
| nucleotide diphosphatase activity |
| phosphodiesterase I activity |
| polysaccharide binding |
| scavenger receptor activity |
| ATP binding |
| protein homodimerization activity |
| 3'-phosphoadenosine 5'-phosphosulfate binding |
| Process |
| metabolic process |
| immune system process |
| immune response |
|
| Cellular Location
|
- Membrane
- Single-pass type II membrane protein
- Single-pass type II membrane protein
- Basolateral cell membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Pantothenate and CoA Biosynthesis |    | 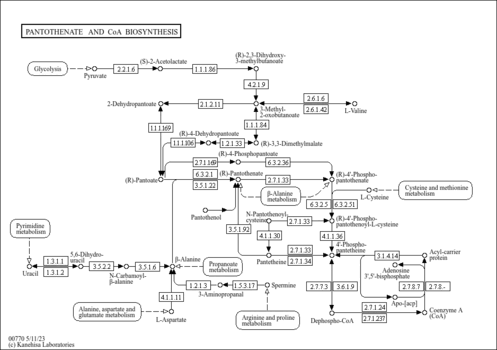 | | Riboflavin Metabolism | 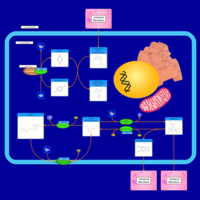 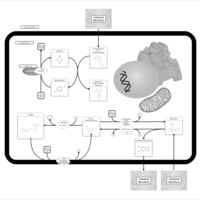 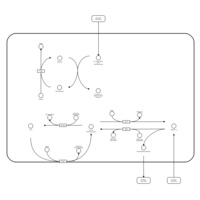 |  | | Nicotinate and Nicotinamide Metabolism | 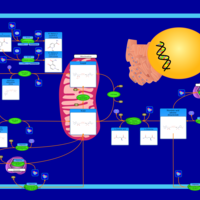 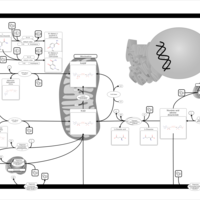 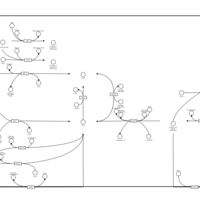 |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 6 |
| Locus
| 6q22-q23 |
| SNPs
| ENPP1 |
| Gene Sequence
|
>2778 bp
ATGGAGCGCGACGGCTGCGCGGGGGGCGGGAGCCGCGGCGGCGAGGGCGGGCGCGCTCCC
CGGGAGGGCCCGGCGGGGAACGGCCGCGATCGGGGCCGCAGCCACGCTGCCGAGGCGCCC
GGGGACCCGCAGGCGGCCGCGTCCTTGCTGGCCCCTATGGACGTGGGGGAGGAGCCGCTG
GAGAAGGCGGCGCGCGCCCGCACTGCCAAGGACCCCAACACCTATAAAGTACTCTCGCTG
GTATTGTCAGTATGTGTGTTAACAACAATACTTGGTTGTATATTTGGGTTGAAACCAAGC
TGTGCCAAAGAAGTTAAAAGTTGCAAAGGTCGCTGTTTCGAGAGAACATTTGGGAACTGT
CGCTGTGATGCTGCCTGTGTTGAGCTTGGAAACTGCTGTTTAGATTACCAGGAGACGTGC
ATAGAACCAGAACATATATGGACTTGCAACAAATTCAGGTGTGGTGAGAAAAGGTTGACC
AGAAGCCTCTGTGCCTGTTCAGATGACTGCAAGGACAAGGGCGACTGCTGCATCAACTAC
AGTTCTGTGTGTCAAGGTGAGAAAAGTTGGGTAGAAGAACCATGTGAGAGCATTAATGAG
CCACAGTGCCCAGCAGGGTTTGAAACGCCTCCTACCCTCTTATTTTCTTTGGATGGATTC
AGGGCAGAATATTTACACACTTGGGGTGGACTTCTTCCTGTTATTAGCAAACTAAAAAAA
TGTGGAACATATACTAAAAACATGAGACCGGTATATCCAACAAAAACTTTCCCCAATCAC
TACAGCATTGTCACCGGATTGTATCCAGAATCTCATGGCATAATCGACAATAAAATGTAT
GATCCCAAAATGAATGCTTCCTTTTCACTTAAAAGTAAAGAGAAATTTAATCCTGAGTGG
TACAAAGGAGAACCAATTTGGGTCACAGCTAAGTATCAAGGCCTCAAGTCTGGCACATTT
TTCTGGCCAGGATCAGATGTGGAAATTAACGGAATTTTCCCAGACATCTATAAAATGTAT
AATGGTTCAGTACCATTTGAAGAAAGGATTTTAGCTGTTCTTCAGTGGCTACAGCTTCCT
AAAGATGAAAGACCACACTTTTACACTCTGTATTTAGAAGAACCAGATTCTTCAGGTCAT
TCATATGGACCAGTCAGCAGTGAAGTCATCAAAGCCTTGCAGAGGGTTGATGGTATGGTT
GGTATGCTGATGGATGGTCTGAAAGAGCTGAACTTGCACAGATGCCTGAACCTCATCCTT
ATTTCAGATCATGGCATGGAACAAGGCAGTTGTAAGAAATACATATATCTGAATAAATAT
TTGGGGGATGTTAAAAATATTAAAGTTATCTATGGACCTGCAGCTCGATTGAGACCCTCT
GATGTCCCAGATAAATACTATTCATTTAACTATGAAGGCATTGCCCGAAATCTTTCTTGC
CGGGAACCAAACCAGCACTTCAAACCTTACCTGAAACATTTCTTACCTAAGCGTTTGCAC
TTTGCTAAGAGTGATAGAATTGAGCCCTTGACATTCTATTTGGACCCTCAGTGGCAACTT
GCATTGAATCCCTCAGAAAGGAAATATTGTGGAAGTGGATTTCATGGCTCTGACAATGTA
TTTTCAAATATGCAAGCCCTCTTTGTTGGCTATGGACCTGGATTCAAGCATGGCATTGAG
GCTGACACCTTTGAAAACATTGAAGTCTATAACTTAATGTGTGATTTACTGAATTTGACA
CCGGCTCCTAATAACGGAACTCATGGAAGTCTTAACCACCTTCTAAAGAATCCTGTTTAT
ACGCCAAAGCATCCCAAAGAAGTGCACCCCCTGGTACAGTGCCCCTTCACAAGAAACCCC
AGAGATAACCTTGGCTGCTCATGTAACCCTTCGATTTTGCCGATTGAGGATTTTCAAACA
CAGTTCAATCTGACTGTGGCAGAAGAGAAGATTATTAAGCATGAAACTTTACCCTATGGA
AGACCTAGAGTTCTCCAGAAGGAAAACACCATCTGTCTTCTTTCCCAGCACCAGTTTATG
AGTGGATACAGCCAAGACATCTTAATGCCCCTTTGGACATCCTATACCGTGGACAGAAAT
GACAGTTTCTCTACGGAAGACTTCTCCAACTGTCTGTACCAGGACTTTAGAATTCCTCTT
AGTCCTGTCCATAAATGTTCATTTTATAAAAATAACACCAAAGTGAGTTACGGGTTCCTC
TCCCCACCACAACTAAATAAAAATTCAAGTGGAATATATTCTGAAGCTTTGCTTACTACA
AATATAGTGCCAATGTACCAGAGTTTTCAAGTTATATGGCGCTACTTTCATGACACCCTA
CTGCGAAAGTATGCTGAAGAAAGAAATGGTGTCAATGTCGTCAGTGGTCCTGTGTTTGAC
TTTGATTATGATGGACGTTGTGATTCCTTAGAGAATCTGAGGCAAAAAAGAAGAGTCATC
CGTAACCAAGAAATTTTGATTCCAACTCACTTCTTTATTGTGCTAACAAGCTGTAAAGAT
ACATCTCAGACGCCTTTGCACTGTGAAAACCTAGACACCTTAGCTTTCATTTTGCCTCAC
AGGACTGATAACAGCGAGAGCTGTGTGCATGGGAAGCATGACTCCTCATGGGTTGAAGAA
TTGTTAATGTTACACAGAGCACGGATCACAGATGTTGAGCACATCACTGGACTCAGCTTC
TATCAACAAAGAAAAGAGCCAGTTTCAGACATTTTAAAGTTGAAAACACATTTGCCAACC
TTTAGCCAAGAAGACTGA
|
| Protein Properties |
|---|
| Number of Residues
| 925 |
| Molecular Weight
| 104923.58 |
| Theoretical pI
| 7.136 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ectonucleotide pyrophosphatase/phosphodiesterase family member 1
MERDGCAGGGSRGGEGGRAPREGPAGNGRDRGRSHAAEAPGDPQAAASLLAPMDVGEEPL
EKAARARTAKDPNTYKVLSLVLSVCVLTTILGCIFGLKPSCAKEVKSCKGRCFERTFGNC
RCDAACVELGNCCLDYQETCIEPEHIWTCNKFRCGEKRLTRSLCACSDDCKDKGDCCINY
SSVCQGEKSWVEEPCESINEPQCPAGFETPPTLLFSLDGFRAEYLHTWGGLLPVISKLKK
CGTYTKNMRPVYPTKTFPNHYSIVTGLYPESHGIIDNKMYDPKMNASFSLKSKEKFNPEW
YKGEPIWVTAKYQGLKSGTFFWPGSDVEINGIFPDIYKMYNGSVPFEERILAVLQWLQLP
KDERPHFYTLYLEEPDSSGHSYGPVSSEVIKALQRVDGMVGMLMDGLKELNLHRCLNLIL
ISDHGMEQGSCKKYIYLNKYLGDVKNIKVIYGPAARLRPSDVPDKYYSFNYEGIARNLSC
REPNQHFKPYLKHFLPKRLHFAKSDRIEPLTFYLDPQWQLALNPSERKYCGSGFHGSDNV
FSNMQALFVGYGPGFKHGIEADTFENIEVYNLMCDLLNLTPAPNNGTHGSLNHLLKNPVY
TPKHPKEVHPLVQCPFTRNPRDNLGCSCNPSILPIEDFQTQFNLTVAEEKIIKHETLPYG
RPRVLQKENTICLLSQHQFMSGYSQDILMPLWTSYTVDRNDSFSTEDFSNCLYQDFRIPL
SPVHKCSFYKNNTKVSYGFLSPPQLNKNSSGIYSEALLTTNIVPMYQSFQVIWRYFHDTL
LRKYAEERNGVNVVSGPVFDFDYDGRCDSLENLRQKRRVIRNQEILIPTHFFIVLTSCKD
TSQTPLHCENLDTLAFILPHRTDNSESCVHGKHDSSWVEELLMLHRARITDVEHITGLSF
YQQRKEPVSDILKLKTHLPTFSQED
|
| External Links |
|---|
| GenBank ID Protein
| 170650661 |
| UniProtKB/Swiss-Prot ID
| P22413 |
| UniProtKB/Swiss-Prot Entry Name
| ENPP1_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_006208.2 |
| GeneCard ID
| ENPP1 |
| GenAtlas ID
| ENPP1 |
| HGNC ID
| HGNC:3356 |
| References |
|---|
| General References
| Not Available |


