| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:37:27 UTC |
|---|
| Updated at | 2020-12-07 19:10:57 UTC |
|---|
| CannabisDB ID | CDB004794 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Cholesterol |
|---|
| Description | Cholesterol, also known as cholesterin or cordulan, belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. Thus, cholesterol is considered to be a sterol lipid molecule. Cholesterol is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Cholesterol is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (3beta,14beta,17alpha)-Cholest-5-en-3-ol | ChEBI | | Cholest-5-en-3beta-ol | ChEBI | | Cholesterin | ChEBI | | (3b,14b,17a)-Cholest-5-en-3-ol | Generator | | (3Β,14β,17α)-cholest-5-en-3-ol | Generator | | Cholest-5-en-3b-ol | Generator | | Cholest-5-en-3β-ol | Generator | | (3Β)-cholest-5-en-3-ol | HMDB | | (3beta)-Cholest-5-en-3-ol | HMDB | | 3Β-hydroxycholest-5-ene | HMDB | | 3beta-Hydroxycholest-5-ene | HMDB | | 5:6-Cholesten-3β-ol | HMDB | | 5:6-Cholesten-3beta-ol | HMDB | | (-)-Cholesterol | HMDB | | Cholesterine | HMDB | | Cholesterol base H | HMDB | | Cholesteryl alcohol | HMDB | | Cholestrin | HMDB | | Cholestrol | HMDB | | Cordulan | HMDB | | Dastar | HMDB | | Dusoline | HMDB | | Dusoran | HMDB | | Dythol | HMDB | | Fancol CH | HMDB | | Hydrocerin | HMDB | | Kathro | HMDB | | Lanol | HMDB | | Super hartolan | HMDB | | Tegolan | HMDB | | Cholesterol | HMDB | | delta5-Cholesten-3beta-ol | PhytoBank | | Δ5-Cholesten-3β-ol | PhytoBank |
|
|---|
| Chemical Formula | C27H46O |
|---|
| Average Molecular Weight | 386.65 |
|---|
| Monoisotopic Molecular Weight | 386.3549 |
|---|
| IUPAC Name | (1S,2R,5S,10S,11S,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-7-en-5-ol |
|---|
| Traditional Name | (1S,2R,5S,10S,11S,14R,15R)-2,15-dimethyl-14-[(2R)-6-methylheptan-2-yl]tetracyclo[8.7.0.0^{2,7}.0^{11,15}]heptadec-7-en-5-ol |
|---|
| CAS Registry Number | 80356-14-5 |
|---|
| SMILES | [H][C@@]1(CC[C@@]2([H])[C@]3([H])CC=C4C[C@@H](O)CC[C@]4(C)[C@@]3([H])CC[C@]12C)[C@H](C)CCCC(C)C |
|---|
| InChI Identifier | InChI=1S/C27H46O/c1-18(2)7-6-8-19(3)23-11-12-24-22-10-9-20-17-21(28)13-15-26(20,4)25(22)14-16-27(23,24)5/h9,18-19,21-25,28H,6-8,10-17H2,1-5H3/t19-,21+,22+,23-,24+,25+,26+,27-/m1/s1 |
|---|
| InChI Key | HVYWMOMLDIMFJA-DPAQBDIFSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as cholesterols and derivatives. Cholesterols and derivatives are compounds containing a 3-hydroxylated cholestane core. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Steroids and steroid derivatives |
|---|
| Sub Class | Cholestane steroids |
|---|
| Direct Parent | Cholesterols and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Cholesterol-skeleton
- Cholesterol
- 3-beta-hydroxysteroid
- 3-beta-hydroxy-delta-5-steroid
- Hydroxysteroid
- 3-hydroxysteroid
- 3-hydroxy-delta-5-steroid
- Delta-5-steroid
- Cyclic alcohol
- Secondary alcohol
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Alcohol
- Aliphatic homopolycyclic compound
|
|---|
| Molecular Framework | Aliphatic homopolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 148 °C | Not Available | | Boiling Point | 360 °C | Wikipedia | | Water Solubility | 9.5e-05 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0a4l-7922000000-36e8a5e1a77d2e71d1d1 | 2014-09-20 | View Spectrum | | GC-MS | Cholesterol, 1 TMS, GC-MS Spectrum | splash10-004i-3911000000-52f5261007adc218f8f8 | Spectrum | | GC-MS | Cholesterol, non-derivatized, GC-MS Spectrum | splash10-052b-2932000000-1667c83d002043a59fff | Spectrum | | GC-MS | Cholesterol, non-derivatized, GC-MS Spectrum | splash10-000l-9527000000-5529a262047f5369c9c1 | Spectrum | | GC-MS | Cholesterol, non-derivatized, GC-MS Spectrum | splash10-0mkr-2954000000-60e7d1f5973e4de33dec | Spectrum | | GC-MS | Cholesterol, non-derivatized, GC-MS Spectrum | splash10-014i-1009000000-e82b8e23dedb45ce70e6 | Spectrum | | GC-MS | Cholesterol, non-derivatized, GC-MS Spectrum | splash10-056v-2900000000-04687c9f19ff52ba4654 | Spectrum | | Predicted GC-MS | Cholesterol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-05i3-1109000000-8cd386b595bd5151c5a2 | Spectrum | | Predicted GC-MS | Cholesterol, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-3104900000-698223f49da0b0c1cf81 | Spectrum | | Predicted GC-MS | Cholesterol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cholesterol, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Cholesterol, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - n/a 27V, positive | splash10-052u-0690000000-36b4aea6637bdf069b5d | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 5V, positive | splash10-014i-3639000000-18a9275beb9bae48c1d7 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 10V, positive | splash10-014j-4934000000-f755ada933cfcf53c303 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 15V, positive | splash10-0002-5921000000-e7d43c825b9f6d45e679 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 20V, positive | splash10-0532-5910000000-ce90c49b7429a03a65da | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 30V, positive | splash10-05nb-8900000000-2e8913fec7126b5b2a62 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 40V, positive | splash10-0apl-9600000000-3096c7a78c3dd2e6a538 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 50V, positive | splash10-067l-9400000000-1b57543b4cd0a05b9b3f | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - QTOF 60V, positive | splash10-0aru-9300000000-45a29b76dfefce3f01ca | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014r-0009000000-dccd68f70545aeac4fab | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05p9-3149000000-7f25daf2b709c7e0d177 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0c00-6269000000-5333d0216e01a3e43367 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-dc853b29b9e884bbb03b | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0009000000-09d9608700564a6fef78 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0ldi-1009000000-96f6e67651380e1c959e | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0009000000-2d9cbc8b5b1fa392fb9b | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0abj-9173000000-9086873e4b65db1ced63 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-8920000000-bd94fa538afb75d04765 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000i-0009000000-c037f2d2f217e99b79ac | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000i-0009000000-7de5154056e69e7319e1 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-0009000000-96c53273291d4857bde4 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Steroidogenesis |    | 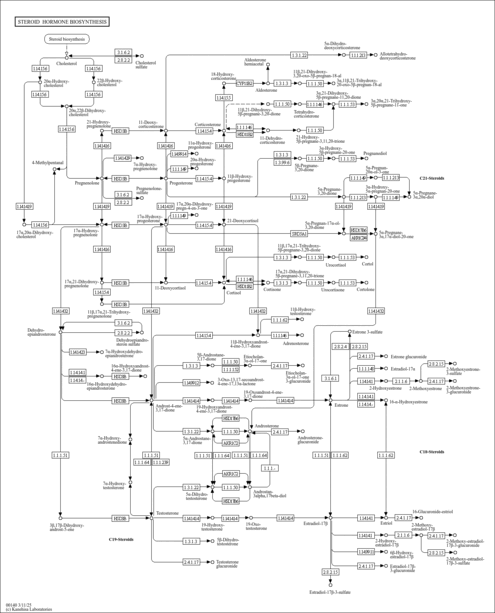 | | Bile Acid Biosynthesis |    | 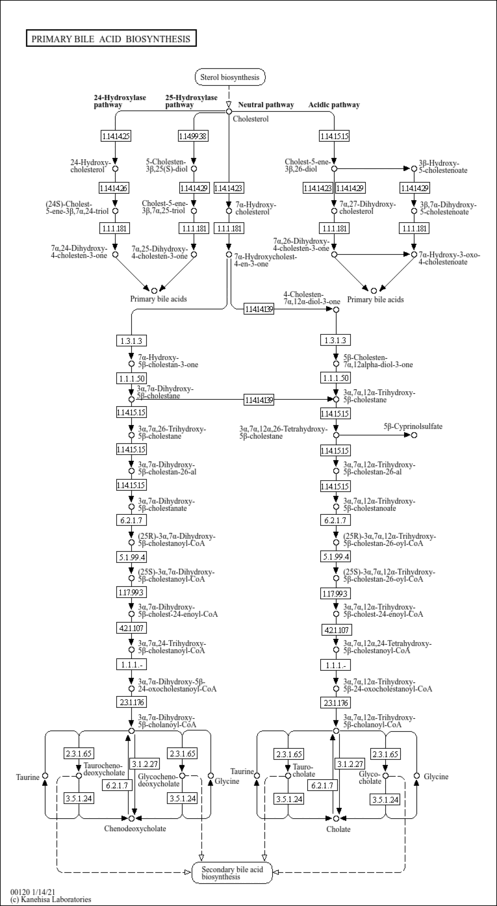 | | Steroid Biosynthesis |  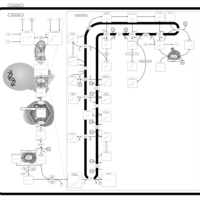 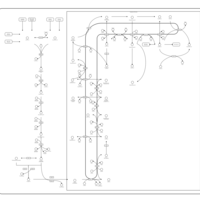 |  | | Smith-Lemli-Opitz Syndrome (SLOS) |  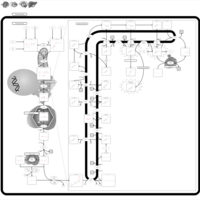 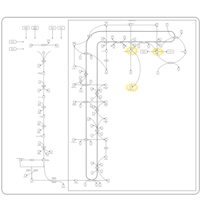 | Not Available | | CHILD Syndrome | 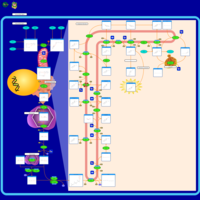 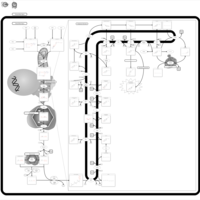 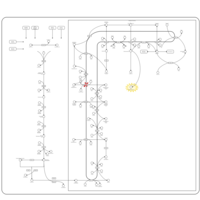 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000067 |
|---|
| DrugBank ID | DB04540 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB013269 |
|---|
| KNApSAcK ID | C00003648 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00187 |
|---|
| BioCyc ID | CHOLESTEROL |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Cholesterol |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 5997 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16113 |
|---|
| References |
|---|
| General References | Not Available |
|---|


