| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:37:21 UTC |
|---|
| Updated at | 2020-12-07 19:10:57 UTC |
|---|
| CannabisDB ID | CDB004793 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Acetoacetic acid |
|---|
| Description | Acetoacetic acid, also known as 3-oxobutanoic acid or 3-oxobutyrate, belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. Acetoacetic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. Acetoacetic acid exists in all living species, ranging from bacteria to humans. Acetoacetic acid is a potentially toxic compound. A 3-oxo monocarboxylic acid that is butyric acid bearing a 3-oxo substituent. Acetoacetic acid, with regard to humans, has been found to be associated with several diseases such as late-onset preeclampsia, crohn's disease, and schizophrenia; acetoacetic acid has also been linked to several inborn metabolic disorders including 2-ketoglutarate dehydrogenase complex deficiency and glucose transporter type 1 deficiency syndrome. Acetoacetic acid is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 3-Ketobutanoic acid | ChEBI | | 3-Ketobutyric acid | ChEBI | | 3-Oxobutanoic acid | ChEBI | | 3-Oxobutyric acid | ChEBI | | beta-Ketobutyric acid | ChEBI | | 3-Ketobutanoate | Generator | | 3-Ketobutyrate | Generator | | 3-Oxobutanoate | Generator | | 3-Oxobutyrate | Generator | | b-Ketobutyrate | Generator | | b-Ketobutyric acid | Generator | | beta-Ketobutyrate | Generator | | Β-ketobutyrate | Generator | | Β-ketobutyric acid | Generator | | Acetoacetate | Generator | | Acetoacetic acid, calcium salt | MeSH | | Acetoacetic acid, lithium salt | MeSH | | Acetoacetic acid, sodium salt | MeSH | | Oxobutyrate | MeSH | | Sodium acetoacetate | MeSH | | 3-oxo-Butanoate | HMDB | | 3-oxo-Butanoic acid | HMDB | | Diacetate | HMDB | | Diacetic acid | HMDB |
|
|---|
| Chemical Formula | C4H6O3 |
|---|
| Average Molecular Weight | 102.09 |
|---|
| Monoisotopic Molecular Weight | 102.0317 |
|---|
| IUPAC Name | 3-oxobutanoic acid |
|---|
| Traditional Name | acetoacetic acid |
|---|
| CAS Registry Number | 541-50-4 |
|---|
| SMILES | CC(=O)CC(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H6O3/c1-3(5)2-4(6)7/h2H2,1H3,(H,6,7) |
|---|
| InChI Key | WDJHALXBUFZDSR-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as short-chain keto acids and derivatives. These are keto acids with an alkyl chain the contains less than 6 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Keto acids and derivatives |
|---|
| Sub Class | Short-chain keto acids and derivatives |
|---|
| Direct Parent | Short-chain keto acids and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Short-chain keto acid
- Beta-keto acid
- 1,3-dicarbonyl compound
- Beta-hydroxy ketone
- Ketone
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 36.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL at 20 °C | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-00kk-3940000000-008eb78d2ba3cfa053b4 | Spectrum | | GC-MS | Acetoacetic acid, 1 MEOX; 1 TMS, GC-MS Spectrum | splash10-000i-9500000000-c3940e591f8aef9d6aac | Spectrum | | GC-MS | Acetoacetic acid, 1 MEOX; 1 TMS, GC-MS Spectrum | splash10-000i-9800000000-67a95675672c3f5447a7 | Spectrum | | GC-MS | Acetoacetic acid, 2 TMS, GC-MS Spectrum | splash10-001i-4950000000-1778836b3908a79b814f | Spectrum | | GC-MS | Acetoacetic acid, 2 TMS, GC-MS Spectrum | splash10-001i-4950000000-4039bb48c98ec17b5c0c | Spectrum | | GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-00kk-3940000000-008eb78d2ba3cfa053b4 | Spectrum | | GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-000i-9500000000-c3940e591f8aef9d6aac | Spectrum | | GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-000i-9800000000-67a95675672c3f5447a7 | Spectrum | | GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-001i-4950000000-1778836b3908a79b814f | Spectrum | | GC-MS | Acetoacetic acid, non-derivatized, GC-MS Spectrum | splash10-001i-4950000000-4039bb48c98ec17b5c0c | Spectrum | | Predicted GC-MS | Acetoacetic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-9f6c0d100736f4d183c9 | Spectrum | | Predicted GC-MS | Acetoacetic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0100-9600000000-470bf0676151478bca3e | Spectrum | | Predicted GC-MS | Acetoacetic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetoacetic acid, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetoacetic acid, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetoacetic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetoacetic acid, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Acetoacetic acid, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, N/A (Annotated) | splash10-000i-9200000000-9b4ab7b6a41632cb125b | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, N/A (Annotated) | splash10-0udi-6900000000-cfab9d4cdb56acafe8cc | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, N/A (Annotated) | splash10-0pb9-9800000000-108aa5ef93455fb1ba28 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , positive | splash10-000i-9100000000-869e2338bbadb32272d4 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 7V, negative | splash10-0a4i-9000000000-3f979a5a82ec557199d2 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-000f-9000000000-2b35e5493fb76f714c22 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-05fr-9000000000-d2734268ae2c1230e50f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-00di-9000000000-0d73b25163774c0d1eb1 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f79-9400000000-081c4a1a05f5581a5ffb | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000l-9100000000-960a06fd90db43d3e0f6 | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-bd091454ccc8f532b42e | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0zfr-9700000000-b01f47bb6cd4f1043983 | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9100000000-d2649829e8a79045e73b | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-1486ce68d51ba83ed499 | 2016-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0pb9-9400000000-2f187d15e3b22b7629d9 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9100000000-1e5e193606848f9d64b4 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-73a6b61cfeb4b8ae9612 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9000000000-de86214ccec455e1860b | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-42af30a03d7e2d4c22fc | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-6aa1f4078505fa3b0971 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Ketone Body Metabolism |    | 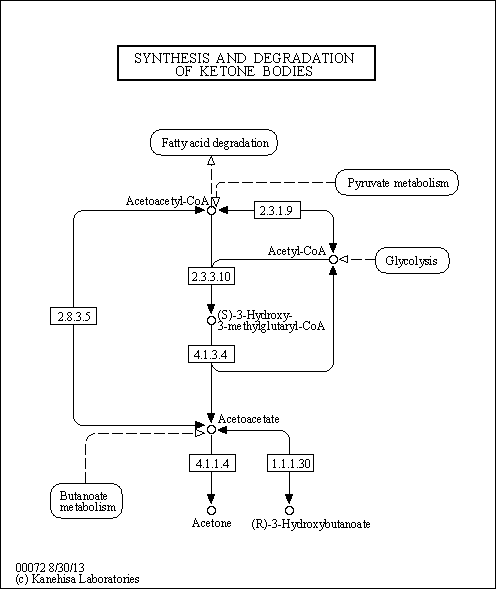 | | Tyrosine Metabolism | 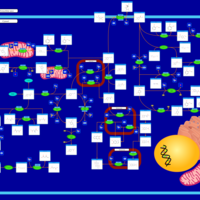 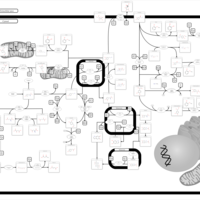 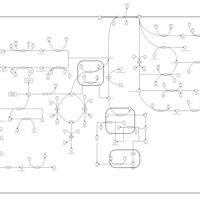 |  | | Butyrate Metabolism | 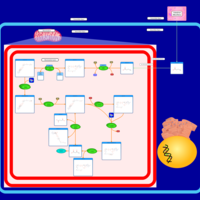 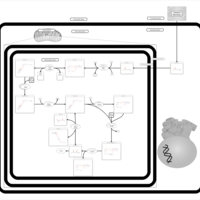 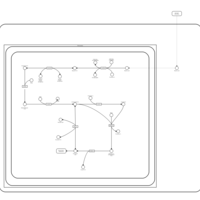 |  | | Valine, Leucine and Isoleucine Degradation |  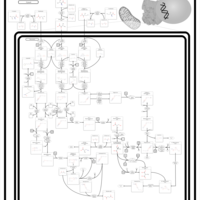 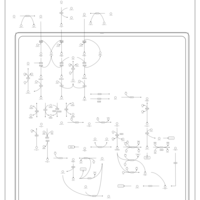 |  | | Phenylalanine and Tyrosine Metabolism | 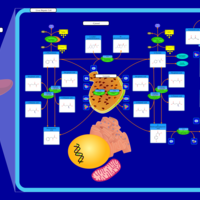 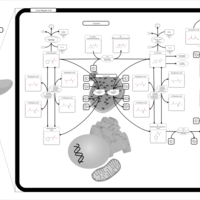 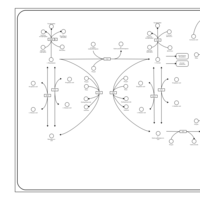 |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Hydroxymethylglutaryl-CoA lyase, mitochondrial | HMGCL | 1p36.1-p35 | P35914 | details | | Aldehyde dehydrogenase, mitochondrial | ALDH2 | 12q24.2 | P05091 | details | | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | BDH1 | 3q29 | Q02338 | details | | Succinyl-CoA:3-ketoacid coenzyme A transferase 1, mitochondrial | OXCT1 | 5p13.1 | P55809 | details | | Fumarylacetoacetase | FAH | 15q25.1 | P16930 | details | | Succinyl-CoA:3-ketoacid coenzyme A transferase 2, mitochondrial | OXCT2 | 1p34 | Q9BYC2 | details | | Hydroxyacid-oxoacid transhydrogenase, mitochondrial | ADHFE1 | 8q13.1 | Q8IWW8 | details | | Acetoacetyl-CoA synthetase | AACS | 12q24.31 | Q86V21 | details | | 3-hydroxymethyl-3-methylglutaryl-CoA lyase, cytoplasmic | HMGCLL1 | 6p12.1 | Q8TB92 | details | | 3-hydroxybutyrate dehydrogenase type 2 | BDH2 | 4q24 | Q9BUT1 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
| Fumarylacetoacetase | FAH | 15q25.1 | P16930 | details | | Hydroxyacid-oxoacid transhydrogenase, mitochondrial | ADHFE1 | 8q13.1 | Q8IWW8 | details | | 3-hydroxymethyl-3-methylglutaryl-CoA lyase, cytoplasmic | HMGCLL1 | 6p12.1 | Q8TB92 | details |
|
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000060 |
|---|
| DrugBank ID | DB01762 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB112157 |
|---|
| KNApSAcK ID | C00007458 |
|---|
| Chemspider ID | 94 |
|---|
| KEGG Compound ID | C00164 |
|---|
| BioCyc ID | 3-KETOBUTYRATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Acetoacetic_acid |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 96 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15344 |
|---|
| References |
|---|
| General References | Not Available |
|---|




