| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:36:44 UTC |
|---|
| Updated at | 2020-12-07 19:10:56 UTC |
|---|
| CannabisDB ID | CDB004787 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Adenine |
|---|
| Description | Adenine, also known as 6-aminopurine or Ade, belongs to the class of organic compounds known as 6-aminopurines. These are purines that carry an amino group at position 6. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. Adenine forms adenosine, a nucleoside, when attached to ribose, and deoxyadenosine when attached to deoxyribose, and it forms adenosine triphosphate (ATP), a nucleotide, when three phosphate groups are added to adenosine. Adenine is a drug which is used for nutritional supplementation, also for treating dietary shortage or imbalance. Adenine is a strong basic compound (based on its pKa). Adenosine triphosphate is used in cellular metabolism as one of the basic methods of transferring chemical energy between reactions. Adenine exists in all living species, ranging from bacteria to humans. In individuals suspected of suffering from a supraventricular tachycardia (SVT), adenosine is used to help identify the rhythm. In humans, adenine is involved in riboflavin metabolism. Adenine is a potentially toxic compound. Adenine (sometimes known as vitamin B4) combines with the sugar ribose to form adenosine, which in turn can be bonded with from one to three phosphoric acid units, yielding AMP, ADP and ATP . A modified form of adenosine monophosphate (cyclic AMP) is an imporant secondary messenger in the propagation of many hormonal stimuli. Adenine is an integral part of the structure of many coenzymes. Adenine is one of four nitrogenous bases utilized in the synthesis of nucleic acids. Certain SVTs can be successfully terminated with adenosine. Adenosine (adenine with a ribose group) causes transient heart block in the AV node of the heart. These adenine derivatives perform important functions in cellular metabolism. Adenine forms adenosine, a nucleoside, when attached to ribose, and deoxyadenosine when attached to deoxyribose, and it forms adenosine triphosphate (ATP), which drives many cellular metabolic processes by transferring chemical energy between reactions. Adenine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 6-Aminopurine | ChEBI | | A | ChEBI | | Ade | ChEBI | | Adenin | ChEBI | | Leucon | Kegg | | 1,6-Dihydro-6-iminopurine | HMDB | | 1H-Purin-6-amine | HMDB | | 1H-Purine-6-amine | HMDB | | 3,6-Dihydro-6-iminopurine | HMDB | | 6-Amino-1H-purine | HMDB | | 6-Amino-3H-purine | HMDB | | 6-Amino-7H-purine | HMDB | | 6-Amino-9H-purine | HMDB | | 6-Amino-purine | HMDB | | 9H-Purin-6-amine | HMDB | | 9H-Purin-6-yl-amin | HMDB | | 9H-Purin-6-ylamine | HMDB | | 9H-Purine-6-amine | HMDB | | Adeninimine | HMDB | | Vitamin b4 | HMDB | | Vitamin b 4 | HMDB | | 4, Vitamin b | HMDB | | b 4, Vitamin | HMDB |
|
|---|
| Chemical Formula | C5H5N5 |
|---|
| Average Molecular Weight | 135.13 |
|---|
| Monoisotopic Molecular Weight | 135.0545 |
|---|
| IUPAC Name | 7H-purin-6-amine |
|---|
| Traditional Name | vitamin B4 |
|---|
| CAS Registry Number | 73-24-5 |
|---|
| SMILES | NC1=C2NC=NC2=NC=N1 |
|---|
| InChI Identifier | InChI=1S/C5H5N5/c6-4-3-5(9-1-7-3)10-2-8-4/h1-2H,(H3,6,7,8,9,10) |
|---|
| InChI Key | GFFGJBXGBJISGV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 6-aminopurines. These are purines that carry an amino group at position 6. Purine is a bicyclic aromatic compound made up of a pyrimidine ring fused to an imidazole ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | 6-aminopurines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 6-aminopurine
- Aminopyrimidine
- Imidolactam
- Pyrimidine
- Heteroaromatic compound
- Imidazole
- Azole
- Azacycle
- Organic nitrogen compound
- Organopnictogen compound
- Hydrocarbon derivative
- Primary amine
- Organonitrogen compound
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 360 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1.03 mg/mL | Not Available | | logP | -0.09 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-000i-6900000000-39944576233751576a91 | 2014-09-20 | View Spectrum | | GC-MS | Adenine, 2 TMS, GC-MS Spectrum | splash10-03di-3490000000-7efe9518c90307a43707 | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-03di-2690000000-6dc072eb8483a2c38e18 | Spectrum | | GC-MS | Adenine, 2 TMS, GC-MS Spectrum | splash10-00di-9350000000-220125189c286547e86c | Spectrum | | GC-MS | Adenine, 2 TMS, GC-MS Spectrum | splash10-03di-4690000000-2d327a6944df53411886 | Spectrum | | GC-MS | Adenine, 1 TMS, GC-MS Spectrum | splash10-0006-3920000000-f488e8aa64272a07b3d9 | Spectrum | | GC-MS | Adenine, 3 TMS, GC-MS Spectrum | splash10-000i-0329000000-0b012fa483ce8764d2af | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-03di-3490000000-7efe9518c90307a43707 | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-03di-2690000000-6dc072eb8483a2c38e18 | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-00di-9350000000-220125189c286547e86c | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-03di-4690000000-2d327a6944df53411886 | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-0006-3920000000-f488e8aa64272a07b3d9 | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-000i-0329000000-0b012fa483ce8764d2af | Spectrum | | GC-MS | Adenine, non-derivatized, GC-MS Spectrum | splash10-03di-2690000000-534edabc8ab24e32f3f5 | Spectrum | | Predicted GC-MS | Adenine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a5i-9700000000-e85c2b0bb19cf3401e5a | Spectrum | | Predicted GC-MS | Adenine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0900000000-b24b09629456779d96e6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0900000000-80808f34c7497219d349 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Negative | splash10-0a5c-6900000000-f6c0abd6d3fca61bd7f7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , negative | splash10-0a4i-0900000000-b24b09629456779d96e6 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-001i-0900000000-80808f34c7497219d349 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a5c-6900000000-f6c0abd6d3fca61bd7f7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-001i-0900000000-cbe0a995dab473351f43 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-001i-0900000000-bfc42d662f222a7c16c7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-4900000000-099ee82fc2f658e1cd04 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-c100f5ac716c497996be | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0900000000-379c5c6ea7b1135677c6 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-014i-9000000000-346f2325bf2e66e7a339 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0900000000-87f60041335919c4a0de | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-014l-9000000000-a71034da1c65738b37bd | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-0a5c-6900000000-f6c0abd6d3fca61bd7f7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-090d369100e8edeb5145 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-001i-0900000000-a56b3bd296195086b5d0 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-a76f7f2350e82c676371 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-053r-0900000000-4213d2fff3cdad674b8e | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-0900000000-d0f2f3ccc46d430b6c3c | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-0900000000-bf20f302482d1cc4a27d | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a59-3900000000-f4155217d6bcdbbcc17f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-066u-9400000000-ada6d909e20e2184a05b | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-0900000000-95d4894082ada0b24773 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-014i-5900000000-66b1c086d7a666b2d02b | 2012-07-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 90 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 50.18 MHz, DMSO-d6, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Purine Metabolism |  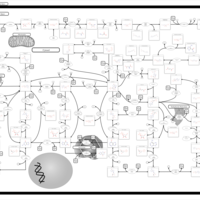  |  | | Adenosine Deaminase Deficiency |  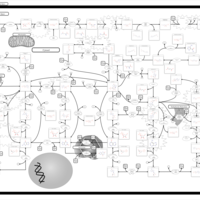  | Not Available | | Adenylosuccinate Lyase Deficiency |    | Not Available | | Gout or Kelley-Seegmiller Syndrome |  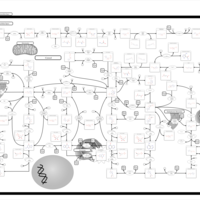 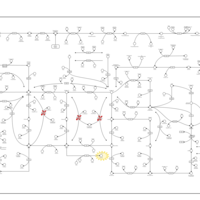 | Not Available | | Lesch-Nyhan Syndrome (LNS) | 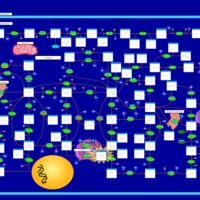 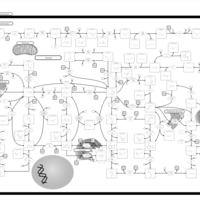 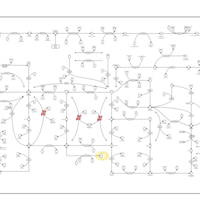 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Equilibrative nucleoside transporter 2 | SLC29A2 | 11q13 | Q14542 | details | | Equilibrative nucleoside transporter 3 | SLC29A3 | 10q22.1 | Q9BZD2 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000034 |
|---|
| DrugBank ID | DB00173 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012266 |
|---|
| KNApSAcK ID | C00001490 |
|---|
| Chemspider ID | 185 |
|---|
| KEGG Compound ID | C00147 |
|---|
| BioCyc ID | ADENINE |
|---|
| BiGG ID | 34039 |
|---|
| Wikipedia Link | Adenine |
|---|
| METLIN ID | 85 |
|---|
| PubChem Compound | 190 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16708 |
|---|
| References |
|---|
| General References | Not Available |
|---|
