| Identification |
|---|
| HMDB Protein ID
| CDBP02367 |
| Secondary Accession Numbers
| Not Available |
| Name
| Purine nucleoside phosphorylase |
| Description
| Not Available |
| Synonyms
|
- Inosine phosphorylase
- PNP
- Inosine-guanosine phosphorylase
|
| Gene Name
| PNP |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in purine-nucleoside phosphorylase activity |
| Specific Function
| The purine nucleoside phosphorylases catalyze the phosphorolytic breakdown of the N-glycosidic bond in the beta-(deoxy)ribonucleoside molecules, with the formation of the corresponding free purine bases and pentose-1-phosphate.
|
| GO Classification
|
| Biological Process |
| purine nucleobase metabolic process |
| purine nucleotide catabolic process |
| positive regulation of T cell proliferation |
| purine-containing compound salvage |
| response to drug |
| immune response |
| inosine catabolic process |
| NAD biosynthesis via nicotinamide riboside salvage pathway |
| nicotinamide riboside catabolic process |
| urate biosynthetic process |
| positive regulation of alpha-beta T cell differentiation |
| interleukin-2 secretion |
| Cellular Component |
| cytoskeleton |
| cytosol |
| Function |
| catalytic activity |
| transferase activity |
| transferase activity, transferring pentosyl groups |
| transferase activity, transferring glycosyl groups |
| purine-nucleoside phosphorylase activity |
| Molecular Function |
| nucleoside binding |
| purine nucleobase binding |
| purine-nucleoside phosphorylase activity |
| drug binding |
| phosphate ion binding |
| Process |
| nucleoside metabolic process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
|
| Cellular Location
|
- Cytoplasm
- cytoskeleton
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | purine nucleoside salvage | Not Available | Not Available | | Purine Metabolism |  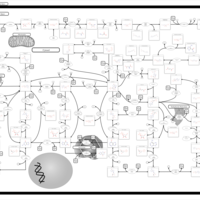 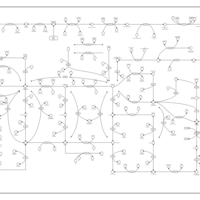 |  | | Adenosine Deaminase Deficiency |  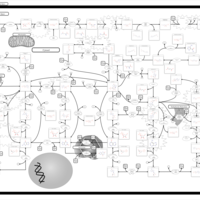 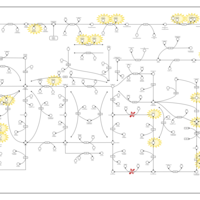 | Not Available | | Adenylosuccinate Lyase Deficiency | 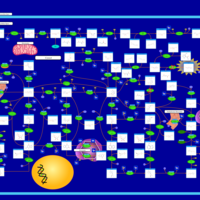 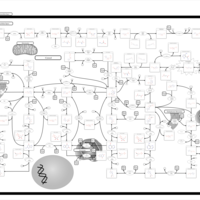 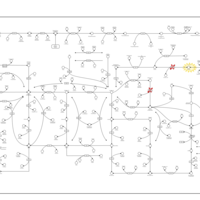 | Not Available | | Gout or Kelley-Seegmiller Syndrome | 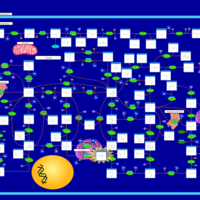 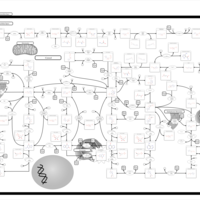  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 14 |
| Locus
| 14q13.1 |
| SNPs
| PNP |
| Gene Sequence
|
>870 bp
ATGGAGAACGGATACACCTATGAAGATTATAAGAACACTGCAGAATGGCTTCTGTCTCAT
ACTAAGCACCGACCTCAAGTTGCAATAATCTGTGGTTCTGGATTAGGAGGTCTGACTGAT
AAATTAACTCAGGCCCAGATCTTTGACTACAGTGAAATCCCCAACTTTCCTCGAAGTACA
GTGCCAGGTCATGCTGGCCGACTGGTGTTTGGGTTCCTGAATGGCAGGGCCTGTGTGATG
ATGCAGGGCAGGTTCCACATGTATGAAGGGTACCCACTCTGGAAGGTGACATTCCCAGTG
AGGGTTTTCCACCTTCTGGGTGTGGACACCCTGGTAGTCACCAATGCAGCAGGAGGGCTG
AACCCCAAGTTTGAGGTTGGAGATATCATGCTGATCCGTGACCATATCAACCTACCTGGT
TTCAGTGGTCAGAACCCTCTCAGAGGGCCCAATGATGAAAGGTTTGGAGATCGTTTCCCT
GCCATGTCTGATGCCTACGACCGGACTATGAGGCAGAGGGCTCTCAGTACCTGGAAACAA
ATGGGGGAGCAACGTGAGCTACAGGAAGGCACCTATGTGATGGTGGCAGGCCCCAGCTTT
GAGACTGTGGCAGAATGTCGTGTGCTGCAGAAGCTGGGAGCAGACGCTGTTGGCATGAGT
ACAGTACCAGAAGTTATCGTTGCACGGCACTGTGGACTTCGAGTCTTTGGCTTCTCACTC
ATCACTAACAAGGTCATCATGGATTATGAAAGCCTGGAGAAGGCCAACCATGAAGAAGTC
TTAGCAGCTGGCAAACAAGCTGCACAGAAATTGGAACAGTTTGTCTCCATTCTTATGGCC
AGCATTCCACTCCCTGACAAAGCCAGTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 289 |
| Molecular Weight
| 32117.69 |
| Theoretical pI
| 6.943 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Purine nucleoside phosphorylase
MENGYTYEDYKNTAEWLLSHTKHRPQVAIICGSGLGGLTDKLTQAQIFDYGEIPNFPRST
VPGHAGRLVFGFLNGRACVMMQGRFHMYEGYPLWKVTFPVRVFHLLGVDTLVVTNAAGGL
NPKFEVGDIMLIRDHINLPGFSGQNPLRGPNDERFGDRFPAMSDAYDRTMRQRALSTWKQ
MGEQRELQEGTYVMVAGPSFETVAECRVLQKLGADAVGMSTVPEVIVARHCGLRVFGFSL
ITNKVIMDYESLEKANHEEVLAAGKQAAQKLEQFVSILMASIPLPDKAS
|
| External Links |
|---|
| GenBank ID Protein
| 35565 |
| UniProtKB/Swiss-Prot ID
| P00491 |
| UniProtKB/Swiss-Prot Entry Name
| PNPH_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| X00737 |
| GeneCard ID
| PNP |
| GenAtlas ID
| PNP |
| HGNC ID
| HGNC:7892 |
| References |
|---|
| General References
| Not Available |
