| Identification |
|---|
| HMDB Protein ID
| CDBP00389 |
| Secondary Accession Numbers
| Not Available |
| Name
| S-methyl-5'-thioadenosine phosphorylase |
| Description
| Not Available |
| Synonyms
|
- 5'-methylthioadenosine phosphorylase
- MTA phosphorylase
- MTAPase
- MTAP
|
| Gene Name
| MTAP |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in transferase activity, transferring pentosyl groups |
| Specific Function
| Catalyzes the reversible phosphorylation of S-methyl-5'-thioadenosine (MTA) to adenine and 5-methylthioribose-1-phosphate. Involved in the breakdown of MTA, a major by-product of polyamine biosynthesis. Responsible for the first step in the methionine salvage pathway after MTA has been generated from S-adenosylmethionine. Has broad substrate specificity with 6-aminopurine nucleosides as preferred substrates.
|
| GO Classification
|
| Biological Process |
| purine ribonucleoside salvage |
| L-methionine salvage from methylthioadenosine |
| polyamine metabolic process |
| nucleobase-containing compound metabolic process |
| Cellular Component |
| cytosol |
| nucleus |
| Function |
| transferase activity, transferring glycosyl groups |
| catalytic activity |
| transferase activity |
| transferase activity, transferring pentosyl groups |
| Molecular Function |
| phosphorylase activity |
| S-methyl-5-thioadenosine phosphorylase activity |
| Process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside metabolic process |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | L-methionine biosynthesis via salvage pathway | Not Available | Not Available | | Cysteine and methionine metabolism | Not Available |  | | Methionine Metabolism | 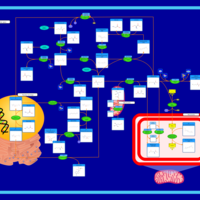 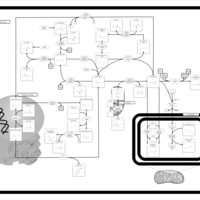 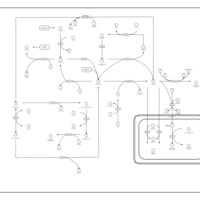 |  | | Cystathionine Beta-Synthase Deficiency | 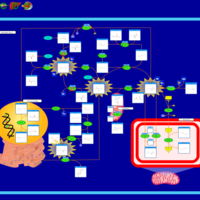   | Not Available | | Hypermethioninemia |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 9 |
| Locus
| 9p21 |
| SNPs
| MTAP |
| Gene Sequence
|
>852 bp
ATGGCCTCTGGCACCACCACCACCGCCGTGAAGATTGGAATAATTGGTGGAACAGGCCTG
GATGATCCAGAAATTTTAGAAGGAAGAACTGAAAAATATGTGGATACTCCATTTGGCAAG
CCATCTGATGCCTTAATTTTGGGGAAGATAAAAAATGTTGATTGCATCCTCCTTGCAAGG
CATGGAAGGCAGCACACCATCATGCCTTCAAAGGTCAACTACCAGGCGAACATCTGGGCT
TTGAAGGAAGAGGGCTGTACACATGTCATAGTGACCACAGCTTGTGGCTCCTTGAGGGAG
GAGATTCAGCCCGGCGATATTGTCATTATTGATCAGTTCATTGACAGGACCACTATGAGA
CCTCAGTCCTTCTATGATGGAAGTCATTCTTGTGCCAGAGGAGTGTGCCATATTCCAATG
GCTGAGCCGTTTTGCCCCAAAACGAGAGAGGTTCTTATAGAGACTGCTAAGAAGCTAGGA
CTCCGGTGCCACTCAAAGGGGACAATGGTCACAATCGAGGGACCTCGTTTTAGCTCCCGG
GCAGAAAGCTTCATGTTCCGCACCTGGGGGGCGGATGTTATCAACATGACCACAGTTCCA
GAGGTGGTTCTTGCTAAGGAGGCTGGAATTTGTTACGCAAGTATCGCCATGGCGACAGAT
TATGACTGCTGGAAGGAGCACGAGGAAGCAGTTTCGGTGGACCGGGTCTTAAAGACCCTG
AAAGAAAACGCTAATAAAGCCAAAAGCTTACTGCTCACTACCATACCTCAGATAGGGTCC
ACAGAATGGTCAGAAACCCTCCATAACCTGAAGAATATGGCCCAGTTTTCTGTTTTATTA
CCAAGACATTAA
|
| Protein Properties |
|---|
| Number of Residues
| 283 |
| Molecular Weight
| 31235.76 |
| Theoretical pI
| 7.188 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>S-methyl-5'-thioadenosine phosphorylase
MASGTTTTAVKIGIIGGTGLDDPEILEGRTEKYVDTPFGKPSDALILGKIKNVDCVLLAR
HGRQHTIMPSKVNYQANIWALKEEGCTHVIVTTACGSLREEIQPGDIVIIDQFIDRTTMR
PQSFYDGSHSCARGVCHIPMAEPFCPKTREVLIETAKKLGLRCHSKGTMVTIEGPRFSSR
AESFMFRTWGADVINMTTVPEVVLAKEAGICYASIAMATDYDCWKEHEEAVSVDRVLKTL
KENANKAKSLLLTTIPQIGSTEWSETLHNLKNMAQFSVLLPRH
|
| External Links |
|---|
| GenBank ID Protein
| 847724 |
| UniProtKB/Swiss-Prot ID
| Q13126 |
| UniProtKB/Swiss-Prot Entry Name
| MTAP_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| U22233 |
| GeneCard ID
| MTAP |
| GenAtlas ID
| MTAP |
| HGNC ID
| HGNC:7413 |
| References |
|---|
| General References
| Not Available |
