| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:26:05 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB000135 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Threonine |
|---|
| Description | Threonine, abbreviated Thr or T, is an essential amino acid in humans. It has a side chain containing a hydroxyl group, making it a polar, uncharged amino acid Threonine is catabolized in mammals largely (70-80%) by threonine dehydrogenase (EC 1.1.1.103) that oxidizes threonine to 2-amino-3-oxobutyrate (which forms glycine and acetyl CoA), and much less by threonine dehydratase (EC 4.2.1.16) that catabolizes threonine into 2-oxobutyrate and ammonia. It is highly concentrated in meat products, cottage cheese and wheat germ. L-Threonine is also found in Cannabis plants (PMID: 6991645 ). It is abundant in human plasma, particularly in newborns. The threonine content of most infant formulas currently on the market is approximately 20% higher than the threonine concentration in human milk. Premature infants fed these formulas have twice the plasma threonine concentrations of breast-fed infants. Increasing the threonine plasma concentrations leads to accumulation of threonine and glycine in the brain. Such accumulation affects the neurotransmitter balance which may have consequences for the brain development during early postnatal life. Thus, excessive threonine intake during infant feeding should be avoided (PMID: 9853925 ). Threonine is an immunostimulant which promotes the growth of thymus gland and probably promotes cell immune defense function. This amino acid has been useful in the treatment of genetic spasticity disorders and multiple sclerosis at a dose of 1 g per day. Severe deficiency of threonine causes neurological dysfunction and lameness in experimental animals |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-Threonine | ChEBI | | (2S,3R)-(-)-Threonine | ChEBI | | (2S,3R)-2-Amino-3-hydroxybutanoic acid | ChEBI | | 2-Amino-3-hydroxybutyric acid | ChEBI | | L-(-)-Threonine | ChEBI | | L-2-Amino-3-hydroxybutyric acid | ChEBI | | L-alpha-Amino-beta-hydroxybutyric acid | ChEBI | | L-Threonin | ChEBI | | T | ChEBI | | Thr | ChEBI | | THREONINE | ChEBI | | (2S,3R)-2-Amino-3-hydroxybutanoate | Generator | | 2-Amino-3-hydroxybutyrate | Generator | | L-2-Amino-3-hydroxybutyrate | Generator | | L-a-Amino-b-hydroxybutyrate | Generator | | L-a-Amino-b-hydroxybutyric acid | Generator | | L-alpha-Amino-beta-hydroxybutyrate | Generator | | L-Α-amino-β-hydroxybutyrate | Generator | | L-Α-amino-β-hydroxybutyric acid | Generator | | (2S,3R)-2-Amino-3-hydroxybutyrate | HMDB | | (2S,3R)-2-Amino-3-hydroxybutyric acid | HMDB | | (R-(R*,s*))-2-amino-3-hydroxybutanoate | HMDB | | (R-(R*,s*))-2-amino-3-hydroxybutanoic acid | HMDB | | (S)-Threonine | HMDB | | 2-Amino-3-hydroxybutanoate | HMDB | | 2-Amino-3-hydroxybutanoic acid | HMDB | | Threonin | HMDB | | [R-(R*,s*)]-2-amino-3-hydroxy-butanoate | HMDB | | [R-(R*,s*)]-2-amino-3-hydroxy-butanoic acid | HMDB | | [R-(R*,s*)]-2-amino-3-hydroxybutanoate | HMDB | | [R-(R*,s*)]-2-amino-3-hydroxybutanoic acid | HMDB | | L Threonine | HMDB |
|
|---|
| Chemical Formula | C4H9NO3 |
|---|
| Average Molecular Weight | 119.12 |
|---|
| Monoisotopic Molecular Weight | 119.0582 |
|---|
| IUPAC Name | (2S,3R)-2-amino-3-hydroxybutanoic acid |
|---|
| Traditional Name | L-threonine |
|---|
| CAS Registry Number | 72-19-5 |
|---|
| SMILES | C[C@@H](O)[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H9NO3/c1-2(6)3(5)4(7)8/h2-3,6H,5H2,1H3,(H,7,8)/t2-,3+/m1/s1 |
|---|
| InChI Key | AYFVYJQAPQTCCC-GBXIJSLDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Beta-hydroxy acid
- Short-chain hydroxy acid
- Hydroxy acid
- Fatty acid
- Amino acid
- Secondary alcohol
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Alcohol
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Primary aliphatic amine
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Organic nitrogen compound
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 256 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 97 mg/mL | Not Available | | logP | -2.94 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Threonine, 3 TMS, GC-MS Spectrum | splash10-0gb9-0930000000-045341234639d940688b | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0gb9-0930000000-08f448150a2533471625 | Spectrum | | GC-MS | L-Threonine, 2 TMS, GC-MS Spectrum | splash10-0159-1910000000-98a39d63665a1a1855dd | Spectrum | | GC-MS | L-Threonine, 3 TMS, GC-MS Spectrum | splash10-014i-1960000000-82e1bd8ff2e302e6b51b | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-d36a7a07f0444e55e38f | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-014i-0980000000-ebba7965f95f804648ab | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0gb9-0930000000-045341234639d940688b | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0gb9-0930000000-08f448150a2533471625 | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0ffa-3921000000-05359581d7eb97866c8f | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-0159-1910000000-98a39d63665a1a1855dd | Spectrum | | GC-MS | L-Threonine, non-derivatized, GC-MS Spectrum | splash10-014i-1960000000-82e1bd8ff2e302e6b51b | Spectrum | | Predicted GC-MS | L-Threonine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00dl-9000000000-a05d200d324c3242c239 | Spectrum | | Predicted GC-MS | L-Threonine, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fdk-8930000000-c4701f28963821b700b3 | Spectrum | | Predicted GC-MS | L-Threonine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TBDMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Threonine, TBDMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-00di-9300000000-85dce837b0f965e73c8a | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4i-9000000000-221510fd551c0b52c362 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4i-9100000000-6ee11649899572cd4867 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-fde7ef1951fddff4b817 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-8900000000-2d7e5609618437e59272 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-4b43567f4a446aed0828 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-014i-4900000000-45382d9abd25be948e5b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-00di-0900000000-071b61d3ea723715c1c1 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a59-9000000000-c8320d0556dbe72049ca | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0uk9-8900000000-9945ecd06408cb733177 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0ufr-8900000000-c9804e2bfc51ec0e8593 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-02t9-0692200000-f96dba3c74726ede32de | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00di-9000000000-94eeca12e76e23c1695e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-0930000000-efff9b10ae39f6ce6095 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-0190000000-74ab3cba7b57b1b1b681 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-014i-1900000000-d9348197a5df9756d30c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-00di-9100000000-9008bb1dfd2806e9f87d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00di-9000000000-47fd01696817c86aecc5 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00di-9000000000-ba9cd5b3e37ded5d3764 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-00di-1900000000-3fa8bd5efd825e5ac3fa | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0kn9-9200000000-063cffd047551fca9ee6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0a4i-9000000000-8b82bffd35f30875a7c0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-0a4i-9000000000-1748dd9a759c98e1c5ee | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-0a4i-9000000000-e9069de219196460cc61 | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uk9-6900000000-d30fcc60bda11aeb88b4 | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 400 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Glycine and Serine Metabolism | 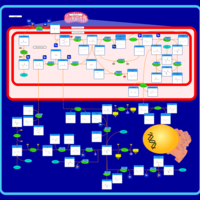 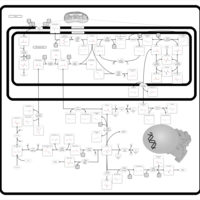 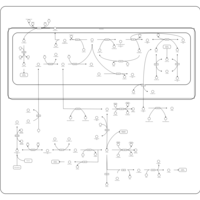 |  | | Threonine and 2-Oxobutanoate Degradation | 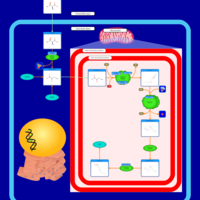 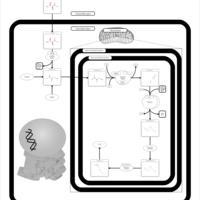 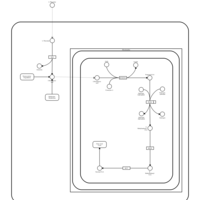 | Not Available | | Amikacin Action Pathway | 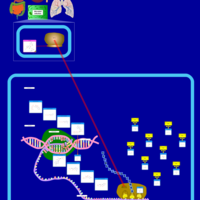 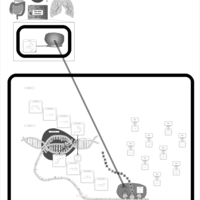  | Not Available | | Azithromycin Action Pathway |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.281 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.241 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.201 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.240 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.129 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.175 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000167 |
|---|
| DrugBank ID | DB00156 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB011999 |
|---|
| KNApSAcK ID | C00001394 |
|---|
| Chemspider ID | 6051 |
|---|
| KEGG Compound ID | C00188 |
|---|
| BioCyc ID | THR |
|---|
| BiGG ID | 34186 |
|---|
| Wikipedia Link | Threonine |
|---|
| METLIN ID | 32 |
|---|
| PubChem Compound | 6288 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16857 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Boehm G, Cervantes H, Georgi G, Jelinek J, Sawatzki G, Wermuth B, Colombo JP: Effect of increasing dietary threonine intakes on amino acid metabolism of the central nervous system and peripheral tissues in growing rats. Pediatr Res. 1998 Dec;44(6):900-6. doi: 10.1203/00006450-199812000-00013. [PubMed:9853925 ]
|
|---|
