| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:24:51 UTC |
|---|
| Updated at | 2020-11-18 16:39:37 UTC |
|---|
| CannabisDB ID | CDB005255 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Mannose 6-phosphate |
|---|
| Description | Mannose 6-phosphate, also known as alpha-D-mannose-6-p or man-6-p, belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. Mannose 6-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). Mannose 6-phosphate exists in all eukaryotes, ranging from yeast to humans. Within humans, mannose 6-phosphate participates in a number of enzymatic reactions. In particular, mannose 6-phosphate can be converted into fructose 6-phosphate through its interaction with the enzyme mannose-6-phosphate isomerase. In addition, mannose 6-phosphate can be biosynthesized from D-mannose; which is mediated by the enzyme hexokinase-1. A D-mannopyranose 6-phosphate with a beta-configuration at the anomeric position. In humans, mannose 6-phosphate is involved in the metabolic disorder called fructosuria. Mannose 6-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Mannose 6-phosphoric acid | Generator | | alpha-D-Mannose-6-p | HMDB | | alpha-D-Mannose-6-phosphate | HMDB | | alpha-delta-Mannose-6-p | HMDB | | alpha-delta-Mannose-6-phosphate | HMDB | | D-Mannose 6-phosphate | HMDB | | delta-Mannose 6-phosphate | HMDB | | Man-6-p | HMDB | | Mannose-6-phosphate | HMDB | | Mannose-6-phosphate dilithium salt | HMDB | | Mannose-6-phosphate disodium salt | HMDB | | Mannose-6-phosphate sodium salt, (D)-isomer | HMDB |
|
|---|
| Chemical Formula | C6H13O9P |
|---|
| Average Molecular Weight | 260.14 |
|---|
| Monoisotopic Molecular Weight | 260.0297 |
|---|
| IUPAC Name | {[(2R,3S,4S,5S,6R)-3,4,5,6-tetrahydroxyoxan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | mannose 6 phosphate |
|---|
| CAS Registry Number | 3672-15-9 |
|---|
| SMILES | O[C@@H]1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h2-10H,1H2,(H2,11,12,13)/t2-,3-,4+,5+,6-/m1/s1 |
|---|
| InChI Key | NBSCHQHZLSJFNQ-RWOPYEJCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexose phosphates. These are carbohydrate derivatives containing a hexose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose phosphate
- Monosaccharide phosphate
- Monoalkyl phosphate
- Organic phosphoric acid derivative
- Alkyl phosphate
- Oxane
- Phosphoric acid ester
- Hemiacetal
- Secondary alcohol
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Alcohol
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Mannose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0005-9740000000-a6e81224fb97751dc2e6 | Spectrum | | Predicted GC-MS | Mannose 6-phosphate, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-2533590000-b6a62e882bfffed3184a | Spectrum | | Predicted GC-MS | Mannose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Mannose 6-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0007-3490000000-734a88a9b3fd55b3e43f | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-052b-9300000000-2c63b28f70b6b2eb75a1 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00l2-9000000000-5406aea843f73d03563c | 2012-07-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0490000000-94ea79ec673f4688b7a0 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dm-5940000000-c649262446626dcb8d6e | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uka-9800000000-aae2cb792906d2dec6cd | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6r-8290000000-8e20219f94b3fe964e38 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-b19b64f5cb4124047baf | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-ecb75a44e3d25affdf31 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a6s-9060000000-b446300429049a22bea4 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-d5aaca13a836cb9f1725 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dm-0970000000-40dbaf3d3c72cf65bf47 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01ot-7900000000-00bc03290b6c0bfb7f84 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000t-9000000000-7a6ff6606f1dbc2f0ae4 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Fructose and Mannose Degradation | 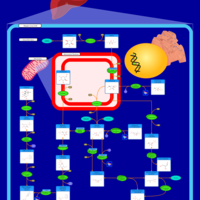 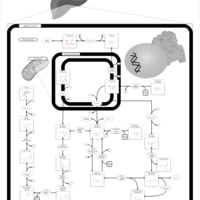 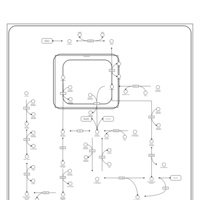 |  | | Fructosuria | 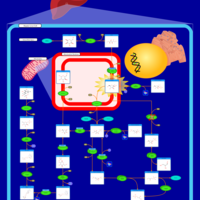   | Not Available | | Fructose intolerance, hereditary |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Cation-dependent mannose-6-phosphate receptor | M6PR | 12p13 | P20645 | details | | Cation-independent mannose-6-phosphate receptor | IGF2R | 6q26 | P11717 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001078 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022411 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 388338 |
|---|
| KEGG Compound ID | C00275 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | 34471 |
|---|
| Wikipedia Link | Mannose 6-phosphate |
|---|
| METLIN ID | 5987 |
|---|
| PubChem Compound | 439198 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 49728 |
|---|
| References |
|---|
| General References | Not Available |
|---|
