| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:10:18 UTC |
|---|
| Updated at | 2021-01-04 18:48:58 UTC |
|---|
| CannabisDB ID | CDB005110 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Hydroquinone |
|---|
| Description | Hydroquinone, also known as quinol or eldoquin, belongs to the class of organic compounds known as hydroquinones. Hydroquinones are compounds containing a hydroquinone moiety, which consists of a benzene ring with a hydroxyl groups at positions 1 and 4 (the para-positions). Hydroquinone is also classified as a phenol. It exists as a white granular solid and is moderately soluble in water. Hydroquinone is produced industrially and used as a water-soluble reducing agent or anti-oxidant. It is a major component in most black and white photographic developer solutions for film and paper where, with the compound metol, it reduces silver halides to elemental silver. Hydroquinone is also widely used as a polymerization inhibitor to prevent radical-initiated polymerization of acrylic acid or methyl acrylate. It is also used as a stabilizing agent in several cosmetics. Medically, hydroquinone has been formulated into creams, lotions and gels as a topical application to whiten skin and reduce skin hyperpigmentation. Over-the-counter sales of creams containing hydroquinone have been limited as it is a potential carcinogen. As a result of these concerns, hydroquinone has been banned in the EU and UK. However, it is still widely used in prescription medications to correct skin discoloration associated with disorders of hyperpigmentation including melasma, post-inflammatory hyperpigmention, sunspots, and freckles. Numerous studies have revealed that hydroquinone can cause exogenous ochronosis, a disfiguring disease in which blue-black pigments are deposited onto the skin, if taken orally; however, skin preparations containing the ingredient are administered topically. Hydroquinone exists in all living organisms, ranging from bacteria to plants to humans. The presence of hydroquinone in normal individuals stems mainly from direct dietary ingestion, catabolism of tyrosine and other substrates by gut bacteria, ingestion of arbutin containing foods, cigarette smoking, and the use of some over-the-counter medicines. Hydroquinone is produced naturally by plants and has been detected in many different plants and plant foods including sweet oranges, anise, bilberries, lingonberries, pears, skunk currants, sesbania flowers, japanese chestnuts, and bamboo shoots. This could make hydroquinone a potential biomarker for the consumption of these foods. Hydroquinone is one of several phenolic compounds found in cannabis smoke and is formed during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,4-Benzenediol | ChEBI | | 1,4-Dihydroxybenzene | ChEBI | | 4-Hydroxyphenol | ChEBI | | Benzene-1,4-diol | ChEBI | | Eldoquin | ChEBI | | p-Benzenediol | ChEBI | | p-Hydroquinone | ChEBI | | p-Hydroxyphenol | ChEBI | | Quinol | ChEBI | | Artra | MeSH | | Eldopaque | MeSH | | Esoterica | MeSH | | Hidroquilaude | MeSH | | Hidroquin | MeSH | | Hidroquinona isdin | MeSH | | Licostrata | MeSH | | Lustra | MeSH | | Melanasa | MeSH | | Melanex | MeSH | | Melpaque | MeSH | | Melquin | MeSH | | Neostrata HQ | MeSH | | Phiaquin | MeSH | | Solaquin | MeSH | | Ultraquin | MeSH | | beta-Quinol | MeSH | | Hydroquinone, copper (1+) salt | MeSH | | Hydroquinone, lead (2+) salt (2:1) | MeSH | | Hydroquinone, monocopper (2+) salt | MeSH | | 1,4-Dihydroxy-benzeen | HMDB | | 1,4-Dihydroxy-benzol | HMDB | | 1,4-Dihydroxybenzen | HMDB | | 1,4-Diidrobenzene | HMDB | | a-Hydroquinone | HMDB | | alpha-Hydroquinone | HMDB | | b-Quinol | HMDB | | Benzohydroquinone | HMDB | | Benzoquinol | HMDB | | Dihydroquinone | HMDB | | Dihydroxybenzene | HMDB | | Hydrochinon | HMDB | | Hydrochinone | HMDB | | Hydroquinol | HMDB | | Hydroquinole | HMDB | | Hydroquinone for synthesis | HMDB | | Hydroquinone GR | HMDB | | Hydroquinoue | HMDB | | Idrochinone | HMDB | | p-Dihydroxybenzene | HMDB | | P-Dioxobenzene | HMDB | | p-Dioxybenzene | HMDB | | P-Hydroxybenzene | HMDB | | Solaquin forte | HMDB | | Eldoquin forte | MeSH, HMDB | | Stratus brand 1 OF hydroquinone | MeSH, HMDB | | ICN brand 1 OF hydroquinone | MeSH, HMDB | | Plough brand 2 OF hydroquinone | MeSH, HMDB | | Eldopaque forte | MeSH, HMDB | | ICN brand 4 OF hydroquinone | MeSH, HMDB | | Black and white | MeSH, HMDB | | ICN brand 2 OF hydroquinone | MeSH, HMDB | | ICN brand 3 OF hydroquinone | MeSH, HMDB | | Plough brand 1 OF hydroquinone | MeSH, HMDB | | Stratus brand 2 OF hydroquinone | MeSH, HMDB | | 1,4-Benzoquinol | PhytoBank | | 1,4-Phenylenediol | PhytoBank | | 1,4-p-Benzenediol | PhytoBank | | p-Dihydroquinone | PhytoBank | | p-Phenylenediol | PhytoBank | | p-Quinol | PhytoBank |
|
|---|
| Chemical Formula | C6H6O2 |
|---|
| Average Molecular Weight | 110.11 |
|---|
| Monoisotopic Molecular Weight | 110.0368 |
|---|
| IUPAC Name | benzene-1,4-diol |
|---|
| Traditional Name | α-hydroquinone |
|---|
| CAS Registry Number | 123-31-9 |
|---|
| SMILES | OC1=CC=C(O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C6H6O2/c7-5-1-2-6(8)4-3-5/h1-4,7-8H |
|---|
| InChI Key | QIGBRXMKCJKVMJ-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hydroquinones. Hydroquinones are compounds containing a hydroquinone moiety, which consists of a benzene ring with a hydroxyl groups at positions 1 and 4. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Phenols |
|---|
| Sub Class | Benzenediols |
|---|
| Direct Parent | Hydroquinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hydroquinone
- 1-hydroxy-2-unsubstituted benzenoid
- Monocyclic benzene moiety
- Organic oxygen compound
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Environmental role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 172.3 °C | Not Available | | Boiling Point | 287 °C | Wikipedia | | Water Solubility | 72 mg/mL at 25 °C | Not Available | | logP | 0.59 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-03di-9500000000-1fa8477944a662535e76 | 2014-09-20 | View Spectrum | | GC-MS | Hydroquinone, 2 TMS, GC-MS Spectrum | splash10-0f79-2490000000-6b4fec222a3499d93790 | Spectrum | | GC-MS | Hydroquinone, non-derivatized, GC-MS Spectrum | splash10-03di-9800000000-c90fd4986fea9691ecbf | Spectrum | | GC-MS | Hydroquinone, non-derivatized, GC-MS Spectrum | splash10-03e9-9300000000-f8b11742557329691e8a | Spectrum | | GC-MS | Hydroquinone, non-derivatized, GC-MS Spectrum | splash10-0f79-2490000000-6b4fec222a3499d93790 | Spectrum | | GC-MS | Hydroquinone, non-derivatized, GC-MS Spectrum | splash10-0f79-2690000000-7306adcc7121a11d67cc | Spectrum | | Predicted GC-MS | Hydroquinone, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03di-5900000000-954dc96d8e3eb85b8324 | Spectrum | | Predicted GC-MS | Hydroquinone, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00e9-8950000000-512c0cf5ddc0b120ec8e | Spectrum | | Predicted GC-MS | Hydroquinone, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Hydroquinone, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-03di-9600000000-e670ba090cb27367db12 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-01p6-9000000000-5fc32c7688c1612df1e7 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000i-9000000000-c5853ce8ef225ea5c353 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (Unknown) , Positive | splash10-03di-9800000000-c90fd4986fea9691ecbf | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-0a4i-0900000000-5635f0157cd37deb1121 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , positive | splash10-03di-2900000000-05be113033a709130eed | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9600000000-e0d15b2e4ea35a7a76be | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-01r6-9000000000-946504408977df40269a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-c16180dd26c127563155 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-03dl-9000000000-cea1cb486f12ea31e3a1 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-01ox-9000000000-3a9d5710c88793e698f1 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-60eeaa79bff371773b2e | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-3a9e293da186985db7fa | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0pb9-9000000000-84bb0a9b4c3cd16da2a7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-7900000000-2256214c81a3e04e01b7 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-8d8328e4f7cd2ebaa27d | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03di-1900000000-9290fcb93f170fc3143f | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01si-9200000000-9f707b9d0c4ad0b73b30 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-d3aae38632ac4c40fd4f | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-74f15490ba4f49cf8c18 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9800000000-1cb01248e88fce387578 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-9f2c82efe2cba7470e17 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-9f2c82efe2cba7470e17 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0f6x-9000000000-8213c13cc381b346f976 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03di-0900000000-fd10fd64675560620b8d | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, DMSO-d6, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 15.08 MHz, DMSO-d6, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Riboflavin Metabolism | 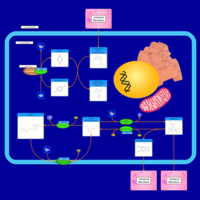 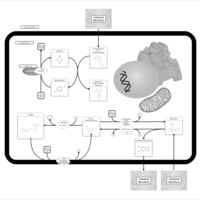 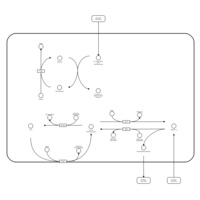 |  | | Glycerolipid Metabolism | 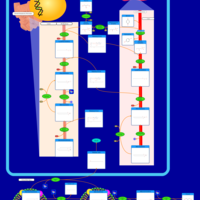 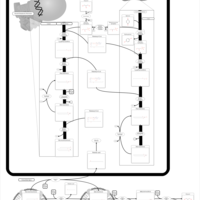 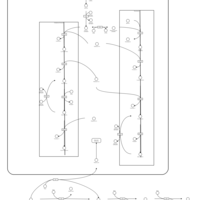 |  | | Glycerol Kinase Deficiency | 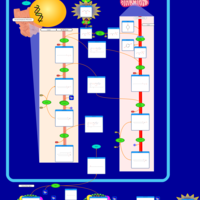 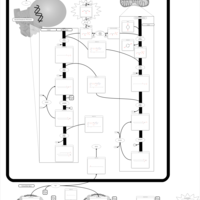  | Not Available | | D-glyceric acidura |    | Not Available | | Familial lipoprotein lipase deficiency | 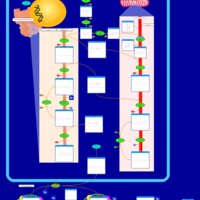  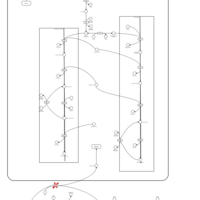 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0002434 |
|---|
| DrugBank ID | DB09526 |
|---|
| Phenol Explorer Compound ID | 660 |
|---|
| FoodDB ID | FDB000885 |
|---|
| KNApSAcK ID | C00002656 |
|---|
| Chemspider ID | 764 |
|---|
| KEGG Compound ID | C00530 |
|---|
| BioCyc ID | HYDROQUINONE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Hydroquinone |
|---|
| METLIN ID | 6681 |
|---|
| PubChem Compound | 785 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17594 |
|---|
| References |
|---|
| General References | Not Available |
|---|

