| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:41:09 UTC |
|---|
| Updated at | 2020-12-07 19:11:03 UTC |
|---|
| CannabisDB ID | CDB004830 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Riboflavin |
|---|
| Description | Riboflavin, also known as lactoflavin or vitamin B2, belongs to the class of organic compounds known as flavins. Flavins are compounds containing a flavin (7,8-dimethyl-benzo[g]pteridine-2,4-dione) moiety, with a structure characterized by an isoalloaxzine tricyclic ring. It occurs in the free form only in the retina of the eye, in whey, and in urine; its principal forms in tissues and cells are as flavin mononucleotide and flavin-adenine dinucleotide. Riboflavin is a drug which is used for the treatment of ariboflavinosis (vitamin b2 deficiency). The antioxidant activity of riboflavin is principally derived from its role as a precursor of FAD and the role of this cofactor in the production of the antioxidant reduced glutathione. Riboflavin is an extremely weak basic (essentially neutral) compound (based on its pKa). Riboflavin exists in all living species, ranging from bacteria to humans. Binds to riboflavin hydrogenase, riboflavin kinase, and riboflavin synthase. In humans, riboflavin is involved in the metabolic disorder called the medium chain acyl-coa dehydrogenase deficiency (mcad) pathway. Outside of the human body, Riboflavin is found, on average, in the highest concentration within a few different foods, such as saffrons, saskatoon berries, and leavening agents and in a lower concentration in grape wines, hippoglossus (common halibut), and spiny lobsters. Riboflavin has also been detected, but not quantified in, several different foods, such as bog bilberries, lowbush blueberries, greenthread tea, nanking cherries, and black radish. This could make riboflavin a potential biomarker for the consumption of these foods. Riboflavin is a potentially toxic compound. Nutritional factor found in milk, eggs, malted barley, liver, kidney, heart, and leafy vegetables. The richest natural source is yeast. Riboflavin is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)pentitol | ChEBI | | 6,7-Dimethyl-9-D-ribitylisoalloxazine | ChEBI | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)benzo[g]pteridine-2,4(3H,10H)-dione | ChEBI | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)isoalloxazine | ChEBI | | 7,8-Dimethyl-10-ribitylisoalloxazine | ChEBI | | e101 | ChEBI | | Lactoflavin | ChEBI | | Riboflavina | ChEBI | | RIBOFLAVINE | ChEBI | | Riboflavinum | ChEBI | | Vitamin b2 | ChEBI | | Vitamin g | ChEBI | | Bisulase | Kegg | | (-)-Riboflavin | HMDB | | 1-Deoxy-1-(3,4-dihydro-7,8-dimethyl-2,4-dioxobenzo[g]pteridin-10(2H)-yl)-D-ribitol | HMDB | | 6,7-Dimethyl-9-ribitylisoalloxazine | HMDB | | 7,8-Dimethyl-10-(D-ribo-2,3,4,5-tetrahydroxypentyl)-benzo[g]pteridine-2,4(3H,10H)-dione | HMDB | | Beflavin | HMDB | | Beflavine | HMDB | | Benzo[g]pteridine riboflavin deriv. | HMDB | | e 101 | HMDB | | Flavaxin | HMDB | | Flavin BB | HMDB | | Flaxain | HMDB | | FOOD Yellow 15 | HMDB | | Hyre | HMDB | | Lactobene | HMDB | | Lactoflavine | HMDB | | Ribipca | HMDB | | Ribocrisina | HMDB | | Riboderm | HMDB | | Ribosyn | HMDB | | Ribotone | HMDB | | Ribovel | HMDB | | Russupteridine yellow III | HMDB | | San yellow b | HMDB | | Vitaflavine | HMDB | | Vitasan b2 | HMDB | | Vitamin b 2 | HMDB |
|
|---|
| Chemical Formula | C17H20N4O6 |
|---|
| Average Molecular Weight | 376.36 |
|---|
| Monoisotopic Molecular Weight | 376.1383 |
|---|
| IUPAC Name | 7,8-dimethyl-10-[(2S,3S,4R)-2,3,4,5-tetrahydroxypentyl]-2H,3H,4H,10H-benzo[g]pteridine-2,4-dione |
|---|
| Traditional Name | riboflavin |
|---|
| CAS Registry Number | 83-88-5 |
|---|
| SMILES | CC1=C(C)C=C2N(C[C@H](O)[C@H](O)[C@H](O)CO)C3=NC(=O)NC(=O)C3=NC2=C1 |
|---|
| InChI Identifier | InChI=1S/C17H20N4O6/c1-7-3-9-10(4-8(7)2)21(5-11(23)14(25)12(24)6-22)15-13(18-9)16(26)20-17(27)19-15/h3-4,11-12,14,22-25H,5-6H2,1-2H3,(H,20,26,27)/t11-,12+,14-/m0/s1 |
|---|
| InChI Key | AUNGANRZJHBGPY-SCRDCRAPSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as flavins. Flavins are compounds containing a flavin (7,8-dimethyl-benzo[g]pteridine-2,4-dione) moiety, with a structure characterized by an isoalloaxzine tricyclic ring. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pteridines and derivatives |
|---|
| Sub Class | Alloxazines and isoalloxazines |
|---|
| Direct Parent | Flavins |
|---|
| Alternative Parents | |
|---|
| Substituents | - Flavin
- Diazanaphthalene
- Quinoxaline
- Pyrimidone
- Pyrazine
- Pyrimidine
- Benzenoid
- Heteroaromatic compound
- Vinylogous amide
- Secondary alcohol
- Lactam
- Polyol
- Azacycle
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 290 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.085 mg/mL | Not Available | | logP | -1.46 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Riboflavin, non-derivatized, GC-MS Spectrum | splash10-006x-4980000000-dd278a577316361d270a | Spectrum | | GC-MS | Riboflavin, non-derivatized, GC-MS Spectrum | splash10-006x-4980000000-dd278a577316361d270a | Spectrum | | Predicted GC-MS | Riboflavin, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0btc-9014000000-75f046dc3c6cb008690e | Spectrum | | Predicted GC-MS | Riboflavin, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0zfs-5146149000-f9db57dd1ccd4a014604 | Spectrum | | Predicted GC-MS | Riboflavin, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_9, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_2_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_3_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_3_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_3_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_3_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Riboflavin, TMS_3_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0a4i-0091000000-a82c54d3153103fcdb1f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0a4i-0091000000-a82c54d3153103fcdb1f | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-2aff124ee1fc62c13844 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-f8b29c3e2c601a944a6c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , negative | splash10-0a4i-0090000000-4e8c9bd38ea0f5ae9a94 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0090000000-5288e9226616bb75603c | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0093000000-5999e10d04a53f4dae9b | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0091000000-8f391045e25f26d53384 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-08fv-0290000000-b4d5f66b726c01d08c55 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0006-4690000000-d195fd5aee22ece7f2ec | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0039000000-549fbc0a59262f64680e | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0195000000-58354696aaddaffc0fd8 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0f76-9231000000-67715e21d6e9ce2655fe | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-0090000000-838afb0228d293bee0a3 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-2190000000-b659be001c9aa265c257 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0a4i-0090000000-21473bfb7f4a3eafd059 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-004i-0019000000-86365dedafa031aa7787 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0006-4390000000-ac1b59ab7cc2209f4241 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dj-4900000000-72d33eb27b9bd6a13d9e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-004l-0569000000-874b71fdc78d04853bf0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-qTof , Positive | splash10-004i-0239000000-659ca9fae9643f3ce73d | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-004l-0569000000-874b71fdc78d04853bf0 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0006-0092000000-1f1be5508c1d50d8dff7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-0006-0092000000-74bf0b86efe72fe37198 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Linear Ion Trap , positive | splash10-057i-0069000000-bb0522be4472e049dbc5 | 2017-09-14 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Riboflavin Metabolism | 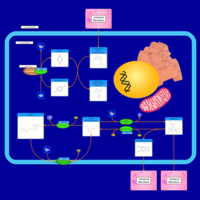 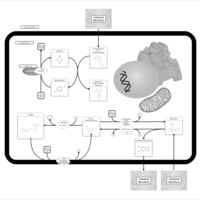  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Solute carrier family 52, riboflavin transporter, member 1 | SLC52A1 | 17p13.2 | Q9NWF4 | details | | Solute carrier family 22 member 6 | SLC22A6 | 11q12.3 | Q4U2R8 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000244 |
|---|
| DrugBank ID | DB00140 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012160 |
|---|
| KNApSAcK ID | C00001552 |
|---|
| Chemspider ID | 431981 |
|---|
| KEGG Compound ID | C00255 |
|---|
| BioCyc ID | RIBOFLAVIN |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Riboflavin |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 493570 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17015 |
|---|
| References |
|---|
| General References | Not Available |
|---|
