| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:40:39 UTC |
|---|
| Updated at | 2020-12-07 19:11:02 UTC |
|---|
| CannabisDB ID | CDB004825 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | O-Phosphoethanolamine |
|---|
| Description | O-Phosphoethanolamine, also known as colamine phosphoric acid or ethanolamine O-phosphate, belongs to the class of organic compounds known as phosphoethanolamines. Phosphoethanolamines are compounds containing a phosphate linked to the second carbon of an ethanolamine. O-Phosphoethanolamine is a very strong basic compound (based on its pKa). O-Phosphoethanolamine exists in all living species, ranging from bacteria to humans. Within humans, O-phosphoethanolamine participates in a number of enzymatic reactions. In particular, O-phosphoethanolamine and palmitaldehyde can be biosynthesized from sphinganine 1-phosphate; which is catalyzed by the enzyme sphingosine-1-phosphate lyase 1. In addition, O-phosphoethanolamine and palmitaldehyde can be biosynthesized from sphingosine 1-phosphate; which is catalyzed by the enzyme sphingosine-1-phosphate lyase 1. In humans, O-phosphoethanolamine is involved in globoid cell leukodystrophy. Outside of the human body, O-Phosphoethanolamine has been detected, but not quantified in, several different foods, such as lambsquarters, nutmegs, yellow wax beans, dandelions, and cloud ear fungus. This could make O-phosphoethanolamine a potential biomarker for the consumption of these foods. The ethanolamine mono-ester of phosphoric acid, and a metabolite of phospholipid metabolism. O-Phosphoethanolamine is a potentially toxic compound. O-Phosphoethanolamine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-Amino-ethanol dihydrogen phosphate | ChEBI | | 2-Amino-ethanol phosphate | ChEBI | | Colamine phosphate | ChEBI | | Colamine phosphoric acid | ChEBI | | Colaminphosphoric acid | ChEBI | | EAP | ChEBI | | Ethanolamine acid phosphate | ChEBI | | Ethanolamine O-phosphate | ChEBI | | Ethanolamine phosphate | ChEBI | | mono(2-Aminoethyl) phosphate | ChEBI | | Monoaminoethyl phosphate | ChEBI | | O-Phosphocolamine | ChEBI | | O-Phosphorylethanolamine | ChEBI | | OPE | ChEBI | | PE | ChEBI | | PEA | ChEBI | | PETN | ChEBI | | Phosphoethanolamine | ChEBI | | Phosphoric acid 2-aminoethyl phenyl ester | ChEBI | | Phosphoryl-ethanolamine | ChEBI | | 2-Amino-ethanol dihydrogen phosphoric acid | Generator | | 2-Amino-ethanol phosphoric acid | Generator | | Colaminphosphate | Generator | | Ethanolamine acid phosphoric acid | Generator | | Ethanolamine O-phosphoric acid | Generator | | Ethanolamine phosphoric acid | Generator | | mono(2-Aminoethyl) phosphoric acid | Generator | | Monoaminoethyl phosphoric acid | Generator | | Phosphate 2-aminoethyl phenyl ester | Generator | | Phosphonoethanolamine | HMDB | | 2-Amino-ethanol dihydrogen phosphate (ester) | HMDB | | 2-Aminoethanol O-phosphate | HMDB | | 2-Aminoethyl dihydrogen phosphate | HMDB | | 2-Aminoethyl dihydrogen phosphate (acd/name 4.0) | HMDB | | 2-Aminoethyl phosphate | HMDB | | Colamine acid phosphate | HMDB | | Colaminephosphoric acid | HMDB | | Ethamp | HMDB | | O-Phosphonatoethanaminium | HMDB | | Phosphoryethanolamine | HMDB | | Phosphorylethanolamine | HMDB | | Phosphorylethanolamine, 3H-labeled CPD | HMDB | | Phosphorylethanolamine magnesium (1:1) salt | HMDB | | Phosphorylethanolamine zinc salt | HMDB | | Phosphorylethanolamine, cobalt (2+) (1:1) salt | HMDB | | Calcium 2-aminoethanol phosphate | HMDB | | Phosphorylethanolamine ca (1:1) salt | HMDB |
|
|---|
| Chemical Formula | C2H8NO4P |
|---|
| Average Molecular Weight | 141.06 |
|---|
| Monoisotopic Molecular Weight | 141.0191 |
|---|
| IUPAC Name | (2-aminoethoxy)phosphonic acid |
|---|
| Traditional Name | phosphorylethanolamine |
|---|
| CAS Registry Number | 1071-23-4 |
|---|
| SMILES | NCCOP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C2H8NO4P/c3-1-2-7-8(4,5)6/h1-3H2,(H2,4,5,6) |
|---|
| InChI Key | SUHOOTKUPISOBE-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as phosphoethanolamines. Phosphoethanolamines are compounds containing a phosphate linked to the second carbon of an ethanolamine. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Organic phosphoric acids and derivatives |
|---|
| Sub Class | Phosphate esters |
|---|
| Direct Parent | Phosphoethanolamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Phosphoethanolamine
- Monoalkyl phosphate
- Alkyl phosphate
- Organic nitrogen compound
- Organic oxygen compound
- Organopnictogen compound
- Organic oxide
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 241 - 243 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 72 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-88dc3b66f275af50de34 | 2014-09-20 | View Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0fki-0910000000-63541b89e6730018e48b | Spectrum | | GC-MS | O-Phosphoethanolamine, 4 TMS, GC-MS Spectrum | splash10-0h9a-1921000000-3a1d4ab89b928259f71d | Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0fki-0910000000-63541b89e6730018e48b | Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0h9a-1921000000-3a1d4ab89b928259f71d | Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0h9a-1921000000-3a1d4ab89b928259f71d | Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0h9a-1921000000-3a1d4ab89b928259f71d | Spectrum | | GC-MS | O-Phosphoethanolamine, non-derivatized, GC-MS Spectrum | splash10-0fki-0911000000-7fc066f17e2ebf6c84f8 | Spectrum | | Predicted GC-MS | O-Phosphoethanolamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-000t-9100000000-543d2d90aa663406e999 | Spectrum | | Predicted GC-MS | O-Phosphoethanolamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0006-9100000000-993b8efd11624567ec4b | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0006-9000000000-f2fd3c258685af07afb5 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0006-9000000000-b479823e7c0ee6b5e03d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0006-2900000000-9cf72ee86018eb78504f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-004i-9000000000-169b03f94a9f53bd0de4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-004i-9000000000-199d7ab7a4aeb040674e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-004i-9000000000-f573b62dada3630598c3 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-004i-9000000000-38c04ce07a85a8db342c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-004i-9100000000-b582c6b51602884756da | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-2900000000-9cf72ee86018eb78504f | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-169b03f94a9f53bd0de4 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-199d7ab7a4aeb040674e | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-f573b62dada3630598c3 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-004i-9000000000-38c04ce07a85a8db342c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-004i-9100000000-b582c6b51602884756da | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-004i-9000000000-4335e927ba2537543f7d | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-004i-9000000000-3e5f10c6e140e555b9fe | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-db65ce46b55b09e54fd2 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-c4e1a40502d86192ed6b | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9300000000-a9eb3cce3bf5c7453bd2 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-4c92e75f13cc6c5119eb | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-5874389316d6cd66936a | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002f-9800000000-b5c7c61b118c38407fa5 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-b562b8507dff78a49e4d | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-dfba6ec4cc15f40c6355 | 2015-04-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 90 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Sphingolipid Metabolism | 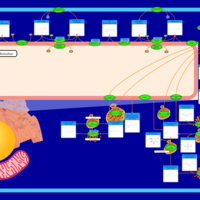 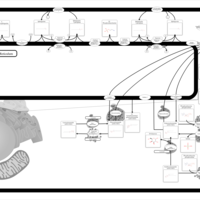 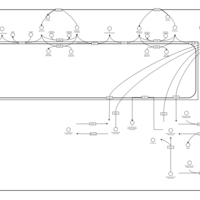 |  | | Gaucher Disease | 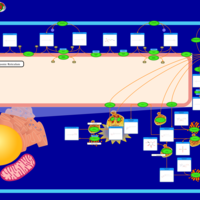 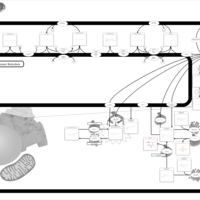 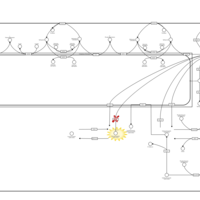 | Not Available | | Globoid Cell Leukodystrophy | 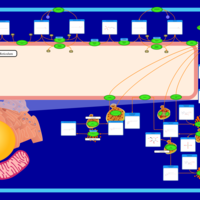 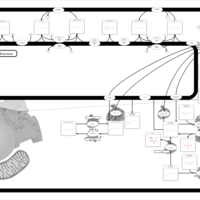 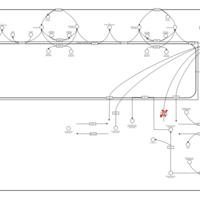 | Not Available | | Metachromatic Leukodystrophy (MLD) | 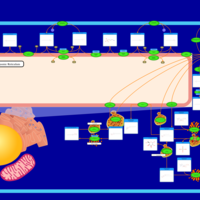 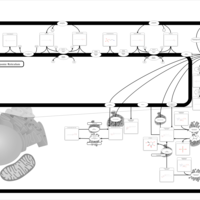 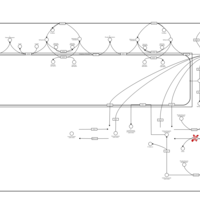 | Not Available | | Fabry disease | 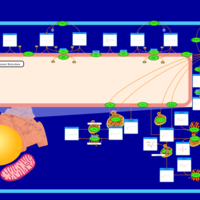 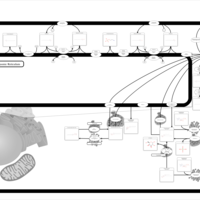 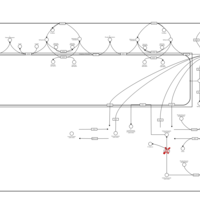 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
| Phosphoethanolamine/phosphocholine phosphatase | PHOSPHO1 | 17q21.32 | Q8TCT1 | details |
|
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000224 |
|---|
| DrugBank ID | DB01738 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB031115 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 990 |
|---|
| KEGG Compound ID | C00346 |
|---|
| BioCyc ID | PHOSPHORYL-ETHANOLAMINE |
|---|
| BiGG ID | 1485304 |
|---|
| Wikipedia Link | Phosphorylethanolamine |
|---|
| METLIN ID | 5233 |
|---|
| PubChem Compound | 1015 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17553 |
|---|
| References |
|---|
| General References | Not Available |
|---|
