| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:39:37 UTC |
|---|
| Updated at | 2020-12-07 19:11:00 UTC |
|---|
| CannabisDB ID | CDB004815 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Dopa |
|---|
| Description | L-Dopa, also known as dopar or dopaston, belongs to the class of organic compounds known as tyrosine and derivatives. Tyrosine and derivatives are compounds containing tyrosine or a derivative thereof resulting from reaction of tyrosine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. L-Dopa is a very strong basic compound (based on its pKa). L-Dopa exists in all living organisms, ranging from bacteria to humans. Within humans, L-dopa participates in a number of enzymatic reactions. In particular, L-dopa and tetrahydrobiopterin can be converted into dopamine and 4a-hydroxytetrahydrobiopterin through its interaction with the enzyme aromatic-L-amino-acid decarboxylase. In addition, L-dopa can be converted into dopaquinone; which is catalyzed by the enzyme tyrosinase. In humans, L-dopa is involved in the metabolic disorder called hawkinsinuria. L-Dopa is an odorless tasting compound. Outside of the human body, L-Dopa is found, on average, in the highest concentration within a few different foods, such as broad beans, swiss chards, and yellow wax beans and in a lower concentration in spinachs, garden onions, and green beans. L-Dopa has also been detected, but not quantified in, several different foods, such as chicory roots, bitter gourds, mustard spinachs, pistachio, and savoy cabbages. This could make L-dopa a potential biomarker for the consumption of these foods. L-Dopa is a potentially toxic compound. L-Dopa, with regard to humans, has been found to be associated with several diseases such as eosinophilic esophagitis and alzheimer's disease; L-dopa has also been linked to several inborn metabolic disorders including aromatic l-amino acid decarboxylase deficiency and sepiapterin reductase deficiency. An optically active form of dopa having L-configuration. L-Dopa is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (-)-3-(3,4-Dihydroxyphenyl)-L-alanine | ChEBI | | (-)-Dopa | ChEBI | | 3,4-Dihydroxy-L-phenylalanine | ChEBI | | 3,4-DIHYDROXYPHENYLALANINE | ChEBI | | 3-Hydroxy-L-tyrosine | ChEBI | | beta-(3,4-Dihydroxyphenyl)-L-alanine | ChEBI | | beta-(3,4-Dihydroxyphenyl)alanine | ChEBI | | Dihydroxy-L-phenylalanine | ChEBI | | Dopar | ChEBI | | L-beta-(3,4-Dihydroxyphenyl)alanine | ChEBI | | Levodopa | ChEBI | | Levodopum | ChEBI | | b-(3,4-Dihydroxyphenyl)-L-alanine | Generator | | Β-(3,4-dihydroxyphenyl)-L-alanine | Generator | | b-(3,4-Dihydroxyphenyl)alanine | Generator | | Β-(3,4-dihydroxyphenyl)alanine | Generator | | L-b-(3,4-Dihydroxyphenyl)alanine | Generator | | L-Β-(3,4-dihydroxyphenyl)alanine | Generator | | (2S)-2-Amino-3-(3,4-dihydroxyphenyl)propanoate | HMDB | | (2S)-2-Amino-3-(3,4-dihydroxyphenyl)propanoic acid | HMDB | | 3,4-Dihydroxyphenyl-L-alanine | HMDB | | 3-(3,4-Dihydroxyphenyl)-L-alanine | HMDB | | b-(3,4-Dihydroxyphenyl)-a-L-alanine | HMDB | | Bendopa | HMDB | | beta-(3,4-Dihydroxyphenyl)-alpha-L-alanine | HMDB | | Cidandopa | HMDB | | Dihydroxyphenylalanine | HMDB | | Dopaflex | HMDB | | Dopaidan | HMDB | | Dopal | HMDB | | Dopalina | HMDB | | Doparkine | HMDB | | Doparl | HMDB | | Dopasol | HMDB | | Dopaston | HMDB | | Dopastone | HMDB | | Dopastral | HMDB | | Dopicar | HMDB | | Doprin | HMDB | | Eldopal | HMDB | | Eldopar | HMDB | | Eldopatec | HMDB | | Eurodopa | HMDB | | Helfo-dopa | HMDB | | Insulamina | HMDB | | L-(-)-Dopa | HMDB | | L-3-(3,4-Dihydroxyphenyl)-alanine | HMDB | | L-4-5-Dihydroxyphenylalanine | HMDB | | L-b-(3,4-Dihydroxyphenyl)-a-alanine | HMDB | | L-beta-(3,4-Dihydroxyphenyl)-alpha-alanine | HMDB | | L-Dihydroxyphenylalanine | HMDB | | Laradopa | HMDB | | Larodopa | HMDB | | Ledopa | HMDB | | Levedopa | HMDB | | Levopa | HMDB | | Maipedopa | HMDB | | Parda | HMDB | | Pardopa | HMDB | | Prodopa | HMDB | | Syndopa | HMDB | | Veldopa | HMDB | | Weldopa | HMDB | | 3 Hydroxy L tyrosine | HMDB | | Roche brand OF levodopa | HMDB | | L 3,4 Dihydroxyphenylalanine | HMDB | | L-3,4-Dihydroxyphenylalanine | HMDB | | Medphano brand OF levodopa | HMDB | | L Dopa | HMDB | | Roberts brand OF levodopa | HMDB |
|
|---|
| Chemical Formula | C9H11NO4 |
|---|
| Average Molecular Weight | 197.19 |
|---|
| Monoisotopic Molecular Weight | 197.0688 |
|---|
| IUPAC Name | (2S)-2-amino-3-(3,4-dihydroxyphenyl)propanoic acid |
|---|
| Traditional Name | levodopa |
|---|
| CAS Registry Number | 59-92-7 |
|---|
| SMILES | N[C@@H](CC1=CC=C(O)C(O)=C1)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C9H11NO4/c10-6(9(13)14)3-5-1-2-7(11)8(12)4-5/h1-2,4,6,11-12H,3,10H2,(H,13,14)/t6-/m0/s1 |
|---|
| InChI Key | WTDRDQBEARUVNC-LURJTMIESA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as tyrosine and derivatives. Tyrosine and derivatives are compounds containing tyrosine or a derivative thereof resulting from reaction of tyrosine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Tyrosine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Tyrosine or derivatives
- Phenylalanine or derivatives
- 3-phenylpropanoic-acid
- Alpha-amino acid
- Amphetamine or derivatives
- L-alpha-amino acid
- Catechol
- 1-hydroxy-4-unsubstituted benzenoid
- Aralkylamine
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Monocyclic benzene moiety
- Benzenoid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Primary amine
- Primary aliphatic amine
- Organic nitrogen compound
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organopnictogen compound
- Amine
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Environmental role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 285 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 5 mg/mL | Not Available | | logP | -2.39 | SANGSTER (1993) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Dopa, 4 TMS, GC-MS Spectrum | splash10-014i-0790000000-b2f7f063a2c8197c7edd | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-0690000000-622497b3104c6082a45d | Spectrum | | GC-MS | L-Dopa, 4 TMS, GC-MS Spectrum | splash10-00xr-9350000000-b0cc4636931d2de64d81 | Spectrum | | GC-MS | L-Dopa, 4 TMS, GC-MS Spectrum | splash10-014i-0590000000-4474e81e4226bb4e1d4c | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-0790000000-b2f7f063a2c8197c7edd | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-0690000000-622497b3104c6082a45d | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-00xr-9350000000-b0cc4636931d2de64d81 | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-0590000000-4474e81e4226bb4e1d4c | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-1890000000-646d209fa1943582a336 | Spectrum | | GC-MS | L-Dopa, non-derivatized, GC-MS Spectrum | splash10-014i-0690000000-720ed87e98a0d9f1721d | Spectrum | | Predicted GC-MS | L-Dopa, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0fk9-3900000000-266d9baeda773fe1fb22 | Spectrum | | Predicted GC-MS | L-Dopa, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-4193000000-8c76bf85d8a897e9403c | Spectrum | | Predicted GC-MS | L-Dopa, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_2_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Dopa, TMS_3_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0uea-0900000000-8eb71aa0cc8622f097a2 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a59-2900000000-bf63b9b719959b82b543 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-056r-9300000000-a78b0b31dd33fe8479a7 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0f6t-0911000000-15affa616923dfb9c45a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-0900000000-22d8267801d0eb0b73c2 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-2c310034a1a871502b4c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0udi-0900000000-5e6020c952f741531fcb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0007-0970100000-49594dae82ce73e734e6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-2183a68f58b951f3f1c7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0900000000-0030db588fbd92c5b761 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0006-0090000000-544615463a975baae9e4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0729111000-0a20b01f58fff8ad7ef0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-c8095a31ed4b3dbbc646 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0190000000-41515cba3a6929721859 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0002-0900000000-8074c509ef5bae1129fc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0006-0502193020-497bfad7ba247159ca00 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0900000000-0b0d4b6dcb7f1fa24e1a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-004i-0029800000-05f40324c8c1fec7963a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0006-0000090000-c0cd80185ce47b30e5fe | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-0002-0900000000-df116b84981cf4a1371a | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) 30V, Positive | splash10-0f6t-0900000000-1c1c39a8880442ea18df | 2012-08-31 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udj-0900000000-cf54b26df05181b0d2fc | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udi-0900000000-d506f2673b114b8e38d2 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-8900000000-1699873cb7650f62b5bc | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-0900000000-a0d81bfc4868b0d1cbf9 | 2017-07-26 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Tyrosine Metabolism | 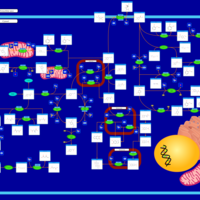 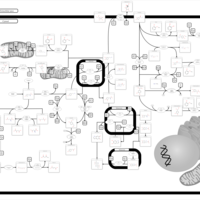 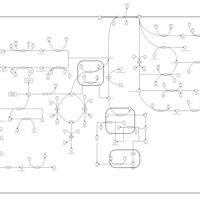 |  | | Catecholamine Biosynthesis | 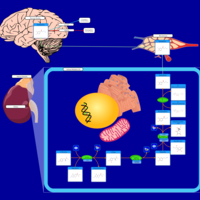 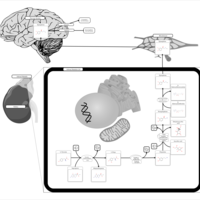 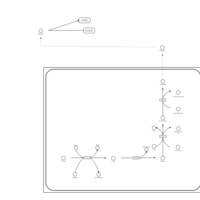 |  | | Aromatic L-Aminoacid Decarboxylase Deficiency | 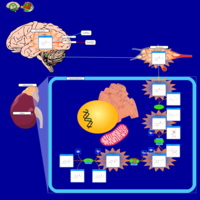 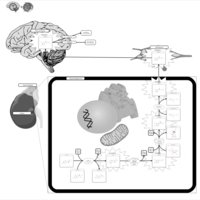 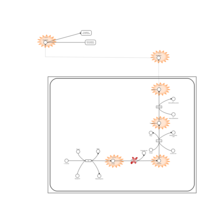 | Not Available | | Tyrosine hydroxylase deficiency | 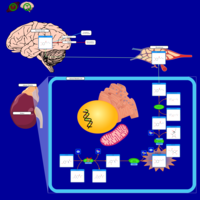 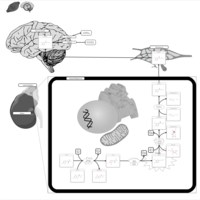 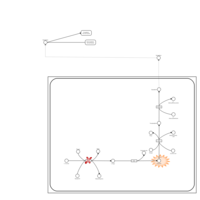 | Not Available | | Alkaptonuria | 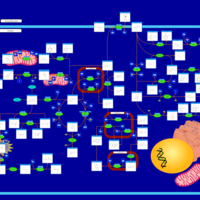 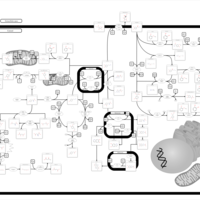 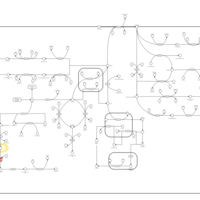 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000181 |
|---|
| DrugBank ID | DB01235 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000567 |
|---|
| KNApSAcK ID | C00001357 |
|---|
| Chemspider ID | 5824 |
|---|
| KEGG Compound ID | C00355 |
|---|
| BioCyc ID | L-DIHYDROXY-PHENYLALANINE |
|---|
| BiGG ID | 34719 |
|---|
| Wikipedia Link | Levodopa |
|---|
| METLIN ID | 42 |
|---|
| PubChem Compound | 6047 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15765 |
|---|
| References |
|---|
| General References | Not Available |
|---|
