| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:15:22 UTC |
|---|
| Updated at | 2020-11-18 16:39:27 UTC |
|---|
| CannabisDB ID | CDB005160 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | D-Erythrose 4-phosphate |
|---|
| Description | D-Erythrose 4-phosphate, also known as D-erythrose-4-p or 4-O-phosphono-D-erythrose, belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. D-Erythrose 4-phosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). D-Erythrose 4-phosphate exists in all living species, ranging from bacteria to humans. Within humans, D-erythrose 4-phosphate participates in a number of enzymatic reactions. In particular, D-erythrose 4-phosphate and fructose 6-phosphate can be converted into D-glyceraldehyde 3-phosphate and D-sedoheptulose 7-phosphate; which is catalyzed by the enzyme transaldolase. In addition, D-erythrose 4-phosphate and xylulose 5-phosphate can be converted into fructose 6-phosphate and D-glyceraldehyde 3-phosphate through its interaction with the enzyme transketolase. In humans, D-erythrose 4-phosphate is involved in the metabolic disorder called the glucose-6-phosphate dehydrogenase deficiency pathway. D-Erythrose 4-phosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-O-Phosphono-D-erythrose | ChEBI | | Erythose-4-phosphate | ChEBI | | D-Erythrose 4-phosphoric acid | Generator | | Erythose-4-phosphoric acid | Generator | | D-Erythrose 4-PO4 | HMDB | | D-Erythrose-4-P | HMDB | | D-Erythrose-4-phosphate | HMDB | | Erythrose 4-phosphate | HMDB, MeSH | | Erythrose 4-PO4 | HMDB | | Erythrose-4-P | HMDB | | Erythrose-4-phosphate | HMDB | | Erythrose-4P | HMDB | | Threose 4-phosphate | HMDB, MeSH | | Erythrose 4-phosphate, (r*,r*)-isomer | MeSH, HMDB | | Erythrose 4-phosphate, ((r*,r*)-(+-))-isomer | MeSH, HMDB | | (2R,3R)-2,3-Dihydroxy-4-(phosphonooxy)butanal | HMDB | | D-Erythrose 4-phosphate | HMDB |
|
|---|
| Chemical Formula | C4H9O7P |
|---|
| Average Molecular Weight | 200.08 |
|---|
| Monoisotopic Molecular Weight | 200.0086 |
|---|
| IUPAC Name | [(2R,3R)-2,3-dihydroxy-4-oxobutoxy]phosphonic acid |
|---|
| Traditional Name | 4-O-phosphono-D-erythrose |
|---|
| CAS Registry Number | 585-18-2 |
|---|
| SMILES | O[C@H](COP(O)(O)=O)[C@@H](O)C=O |
|---|
| InChI Identifier | InChI=1S/C4H9O7P/c5-1-3(6)4(7)2-11-12(8,9)10/h1,3-4,6-7H,2H2,(H2,8,9,10)/t3-,4+/m0/s1 |
|---|
| InChI Key | NGHMDNPXVRFFGS-IUYQGCFVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Monosaccharide phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monosaccharide phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Beta-hydroxy aldehyde
- Alpha-hydroxyaldehyde
- Secondary alcohol
- 1,2-diol
- Organic oxide
- Hydrocarbon derivative
- Carbonyl group
- Aldehyde
- Alcohol
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | D-Erythrose 4-phosphate, 1 MEOX; 4 TMS, GC-MS Spectrum | splash10-0a4i-2946000000-9443f2e925fd6a35c72c | Spectrum | | GC-MS | D-Erythrose 4-phosphate, 1 MEOX; 4 TMS, GC-MS Spectrum | splash10-0a4i-2967000000-d813d71e392e180e1a1a | Spectrum | | GC-MS | D-Erythrose 4-phosphate, non-derivatized, GC-MS Spectrum | splash10-0a4i-2946000000-9443f2e925fd6a35c72c | Spectrum | | GC-MS | D-Erythrose 4-phosphate, non-derivatized, GC-MS Spectrum | splash10-0a4i-2967000000-d813d71e392e180e1a1a | Spectrum | | GC-MS | D-Erythrose 4-phosphate, non-derivatized, GC-MS Spectrum | splash10-0pba-1933000000-ce3489351efc49bf69f3 | Spectrum | | GC-MS | D-Erythrose 4-phosphate, non-derivatized, GC-MS Spectrum | splash10-0ka2-2923000000-16fa49e06a727e2d6a47 | Spectrum | | Predicted GC-MS | D-Erythrose 4-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9600000000-9d57877fd2f223b326b3 | Spectrum | | Predicted GC-MS | D-Erythrose 4-phosphate, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-01di-8973000000-4793f4bb7564afb5ca7b | Spectrum | | Predicted GC-MS | D-Erythrose 4-phosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-002b-9000000000-06d71bced6df9b8baad3 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 14V, positive | splash10-0f89-0069000000-8e4338affc4361b89493 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 14V, positive | splash10-0uk9-0690000000-30a248b95eee8bfd3014 | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0ue9-3940000000-b2cf1408c63fcd508190 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-114m-9800000000-ca839aa0d4b5ed1d26a8 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052v-9100000000-ce3e3be16de64b5ce20e | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-054k-8900000000-951c8d2dad21e2f2c479 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9100000000-4106e1504853e25f6dff | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-150345a1ab7b74a502c9 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0002-9010000000-1278b7bf699cdfb2fc5e | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-db5a64d3322c49bfb840 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-bab6e7f750d40a012094 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002b-9000000000-4212adf2abc696a02cdb | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-c47fda1609169ecd31ba | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-a5e502a2627af2048a1f | 2021-09-25 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Pentose Phosphate Pathway | 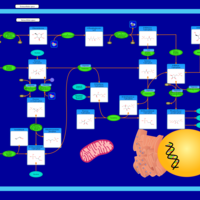  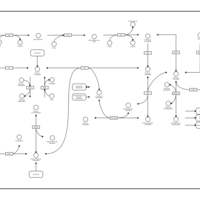 |  | | Glucose-6-phosphate dehydrogenase deficiency | 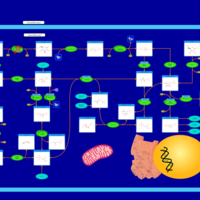 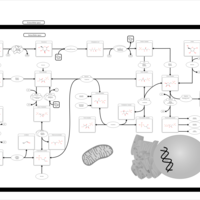 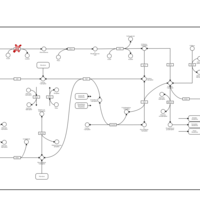 | Not Available | | Ribose-5-phosphate isomerase deficiency | 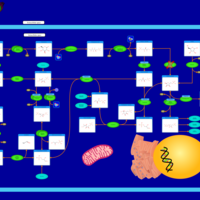 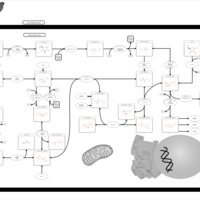 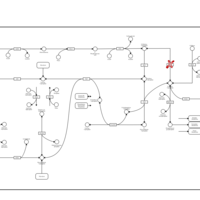 | Not Available | | Transaldolase deficiency | 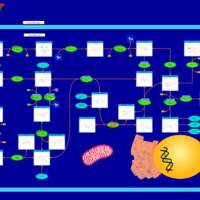 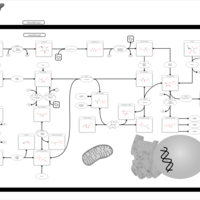 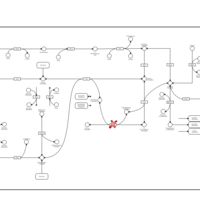 | Not Available | | Warburg Effect | 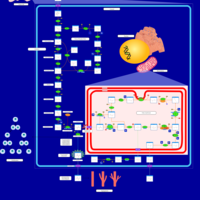 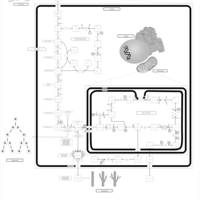 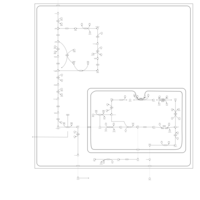 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001321 |
|---|
| DrugBank ID | DB03937 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001614 |
|---|
| KNApSAcK ID | C00007472 |
|---|
| Chemspider ID | 109096 |
|---|
| KEGG Compound ID | C00279 |
|---|
| BioCyc ID | ERYTHROSE-4P |
|---|
| BiGG ID | 34479 |
|---|
| Wikipedia Link | Erythrose_4-phosphate |
|---|
| METLIN ID | 6158 |
|---|
| PubChem Compound | 122357 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 48153 |
|---|
| References |
|---|
| General References | Not Available |
|---|
