| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:11:25 UTC |
|---|
| Updated at | 2022-12-13 23:36:22 UTC |
|---|
| CannabisDB ID | CDB005121 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Histamine |
|---|
| Description | Histamine, also known as histaminum or eramin, belongs to the class of organic compounds known as 2-arylethylamines. These are primary amines that have the general formula RCCNH2, where R is an organic group. Histamine is a very strong basic compound (based on its pKa). Histamine exists in all living species, ranging from bacteria to humans. This can lead to three clinical responses:sneezing due to histamine-associated sensory neural stimulationhyper-secretion from glandular tissuenasal congestion due to vascular engorgement associated with vasodilation and increased capillary permeability Sleep-wake regulation Histamine is a neurotransmitter that is released from histaminergic neurons which project out of the mammalian hypothalamus. Within humans, histamine participates in a number of enzymatic reactions. In particular, histamine can be converted into histamine; which is catalyzed by the enzyme histamine H2 receptor. In addition, histamine can be converted into histamine; which is catalyzed by the enzyme histamine H2 receptor. In humans, histamine is involved in intracellular signalling through histamine H2 receptor and histamine. Histamine is a potentially toxic compound. Histamine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1H-Imidazole-4-ethanamine | ChEBI | | 2-(4-Imidazolyl)ethylamine | ChEBI | | 2-(1H-Imidazol-4-yl)ethanamine | HMDB | | 2-(1H-Imidazol-4-yl)ethylamine | HMDB | | 2-(1H-Imidazol-5-yl)ethanamine | HMDB | | 2-(1H-Imidazol-5-yl)ethylamine | HMDB | | 2-(4-Imidazolyl)ethanamine | HMDB | | 2-Imidazol-4-yl-ethylamine | HMDB | | 2-Imidazol-4-ylethylamine | HMDB | | 4-(2-Aminoethyl)-1H-imidazole | HMDB | | 4-(2-Aminoethyl)imidazole | HMDB | | 4-Imidazoleethylamine | HMDB | | 5-Imidazoleethylamine | HMDB | | b-Imidazolyl-4-ethylamine | HMDB | | beta-Aminoethylglyoxaline | HMDB | | beta-Aminoethylimidazole | HMDB | | beta-Aminothethylglyoxaline | HMDB | | beta-Imidazolyl-4-ethylamine | HMDB | | Eramin | HMDB | | Ergamine | HMDB | | Ergotidine | HMDB | | HSM | HMDB | | 2-(3H-Imidazol-4-yl)ethylamine | HMDB | | Imidazole-4-ethylamine | HMDB | | Β-imidazolyl-4-ethylamine | HMDB | | Histamine | HMDB |
|
|---|
| Chemical Formula | C5H9N3 |
|---|
| Average Molecular Weight | 111.15 |
|---|
| Monoisotopic Molecular Weight | 111.0796 |
|---|
| IUPAC Name | 2-(1H-imidazol-4-yl)ethan-1-amine |
|---|
| Traditional Name | histamine |
|---|
| CAS Registry Number | 51-45-6 |
|---|
| SMILES | NCCC1=CNC=N1 |
|---|
| InChI Identifier | InChI=1S/C5H9N3/c6-2-1-5-3-7-4-8-5/h3-4H,1-2,6H2,(H,7,8) |
|---|
| InChI Key | NTYJJOPFIAHURM-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 2-arylethylamines. These are primary amines that have the general formula RCCNH2, where R is an organic group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Amines |
|---|
| Direct Parent | 2-arylethylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - 2-arylethylamine
- Aralkylamine
- Heteroaromatic compound
- Imidazole
- Azole
- Azacycle
- Organoheterocyclic compound
- Organopnictogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aromatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: Industrial application: Environmental role: |
|---|
| Physical Properties |
|---|
| State | Not Available |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 86 °C | Not Available | | Boiling Point | 209.5 °C | Wikipedia | | Water Solubility | Not Available | Not Available | | logP | -0.70 | SANGSTER (1993) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-9d57c893be8c75ce8178 | 2014-09-20 | View Spectrum | | GC-MS | Histamine, 3 TMS, GC-MS Spectrum | splash10-0f79-5910000000-9946e1707fcaf5562f88 | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-00di-2910000000-8c243055df15d6ce8116 | Spectrum | | GC-MS | Histamine, 3 TMS, GC-MS Spectrum | splash10-00di-9710000000-bf79b524c104e1cc94ba | Spectrum | | GC-MS | Histamine, 2 TMS, GC-MS Spectrum | splash10-0udi-2900000000-a40a3d0aed04ccfe9365 | Spectrum | | GC-MS | Histamine, 3 TMS, GC-MS Spectrum | splash10-00dr-3910000000-44d8fb649d4e75eba66c | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-0f79-5910000000-9946e1707fcaf5562f88 | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-00di-2910000000-8c243055df15d6ce8116 | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-00di-9710000000-bf79b524c104e1cc94ba | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-0udi-2900000000-a40a3d0aed04ccfe9365 | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-00dr-3910000000-44d8fb649d4e75eba66c | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-0udi-2900000000-a40a3d0aed04ccfe9365 | Spectrum | | GC-MS | Histamine, non-derivatized, GC-MS Spectrum | splash10-00dr-3910000000-44d8fb649d4e75eba66c | Spectrum | | Predicted GC-MS | Histamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-9100000000-3de6a14d8a45af6c9d66 | Spectrum | | Predicted GC-MS | Histamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-01ot-9600000000-06c58d75770dfd76aeab | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00kf-9000000000-0135b8f0628e592abc22 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000x-9000000000-6fccc177582b320a48ae | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-03di-1900000000-6cda8885da689473fb42 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-0002-9100000000-37450d9969c24ad44ad7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-0002-9000000000-e9e368926d3146437e13 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-015a-9000000000-f51ad8a8259595600938 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-00lu-9000000000-3e02d5b58a7c9f77ec53 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-03di-0900000000-046407320168835599f0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-03di-3900000000-29ccabc0b6f5396168f2 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-03di-1900000000-6cda8885da689473fb42 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-9100000000-37450d9969c24ad44ad7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-9000000000-e9e368926d3146437e13 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-015a-9000000000-f51ad8a8259595600938 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-00lu-9000000000-77394d92ed9170d75b98 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , positive | splash10-03di-0900000000-046407320168835599f0 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , positive | splash10-01ot-9400000000-a19fc5e555afd53dafe7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9000000000-982b635075f56f26d216 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0frx-9000000000-262a5d1ecfe104fb5771 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03dj-9700000000-b06ddb1eb7ed3ddf4f69 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9200000000-a93565f24c40eddb6cf4 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0uxr-9000000000-71d70766bb4f9d3b13a0 | 2015-04-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-03di-2900000000-2705dc2d177edbc4faa4 | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-03di-6900000000-9e9ebaae6f68b4c2d89d | 2015-04-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9000000000-9d4fe74f18b21423615d | 2015-04-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 90 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 50.18 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Histidine Metabolism | 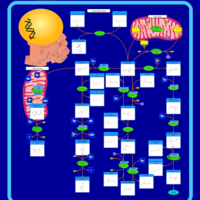 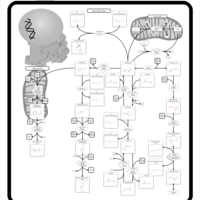 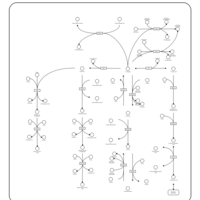 |  | | Histidinemia | 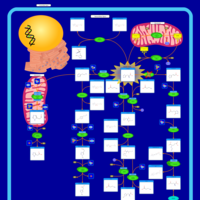  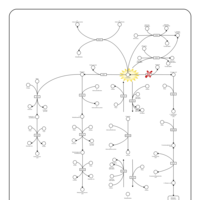 | Not Available | | Gastric Acid Production | 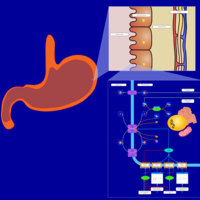 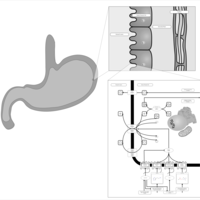  | Not Available | | Esomeprazole Action Pathway | 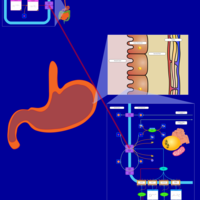 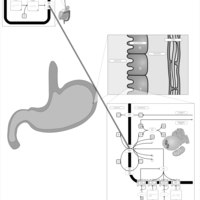  | Not Available | | Omeprazole Action Pathway | 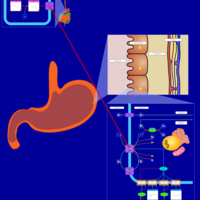   | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 0.000577 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 0.000297 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 0.000904 mg/g dry wt | | details | | Quadra | Detected and Quantified | 0.000557 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 0.000165 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 0.000256 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000870 |
|---|
| DrugBank ID | DB05381 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012596 |
|---|
| KNApSAcK ID | C00001414 |
|---|
| Chemspider ID | 753 |
|---|
| KEGG Compound ID | C00388 |
|---|
| BioCyc ID | HISTAMINE |
|---|
| BiGG ID | 1810403 |
|---|
| Wikipedia Link | Histamine |
|---|
| METLIN ID | 68 |
|---|
| PubChem Compound | 774 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 18295 |
|---|
| References |
|---|
| General References | Not Available |
|---|
