| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:55:28 UTC |
|---|
| Updated at | 2020-11-18 16:39:07 UTC |
|---|
| CannabisDB ID | CDB004967 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Selenomethionine |
|---|
| Description | Selenomethionine, also known as sethotope, belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. Selenomethionine is a very strong basic compound (based on its pKa). selenomethionine can be converted into se-adenosylselenomethionine and phosphoric acid through its interaction with the enzyme S-adenosylmethionine synthase. In humans, selenomethionine is involved in selenoamino acid metabolism. The L-enantiomer of selenomethionine. Selenomethionine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Selenomethionine, (+,-)-isomer | HMDB | | Selenomethionine se 75 | HMDB | | Radioselenomethionine | HMDB | | Selenomethionine, (S)-isomer | HMDB | | Sethotope | HMDB | | Se 75, selenomethionine | HMDB | | Selenomethionine hydrochloride, (S)-isomer | HMDB | | (2S)-2-Amino-4-(methylseleno)butanoic acid | HMDB | | (2S)-2-Azaniumyl-4-methylselanylbutanoate | HMDB | | L-Selenomethionine | HMDB | | Seleno-L-methionine | HMDB | | 2-Amino-4-(methylseleno)butanoic acid | HMDB | | 2-Amino-4-(methylseleno)butyric acid | HMDB | | 2-Amino-4-(methylselenyl)butyric acid | HMDB | | Selenium methionine | HMDB | | Selenomethionine | MeSH |
|
|---|
| Chemical Formula | C5H11NO2Se |
|---|
| Average Molecular Weight | 196.11 |
|---|
| Monoisotopic Molecular Weight | 196.9955 |
|---|
| IUPAC Name | (2S)-2-amino-4-(methylselanyl)butanoic acid |
|---|
| Traditional Name | L-selenomethionine |
|---|
| CAS Registry Number | 3211-76-5 |
|---|
| SMILES | C[Se]CC[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C5H11NO2Se/c1-9-3-2-4(6)5(7)8/h4H,2-3,6H2,1H3,(H,7,8)/t4-/m0/s1 |
|---|
| InChI Key | RJFAYQIBOAGBLC-BYPYZUCNSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Fatty acid
- Amino acid
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Selenoether
- Amine
- Hydrocarbon derivative
- Organic oxide
- Organopnictogen compound
- Primary amine
- Organoselenium compound
- Organooxygen compound
- Organonitrogen compound
- Organic oxygen compound
- Primary aliphatic amine
- Organic nitrogen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 275 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Selenomethionine, 1 TMS, GC-MS Spectrum | splash10-0pc0-2910000000-c7140386a1899064bb79 | Spectrum | | GC-MS | Selenomethionine, 2 TMS, GC-MS Spectrum | splash10-00b9-0950000000-05c9f3aa6ccd78087856 | Spectrum | | Predicted GC-MS | Selenomethionine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0f89-0900000000-4949659ad5c41236a070 | 2020-03-10 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4i-9500000000-dbac3b4e6e034ebd3855 | 2020-03-10 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4i-9200000000-d58c983bc06091b8dd74 | 2020-03-10 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , positive | splash10-0kai-2900000000-86628c3188ed75c0a1e6 | 2020-03-10 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0006-9000000000-177c9b324903fd3e8d24 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0kai-2900000000-c78bec4d8caa8d16c051 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9700000000-20cf34caac47550586a1 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9500000000-76fe5651b983cac7009a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0kai-2900000000-4a553db248ff377f5b2d | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9600000000-8cd75528c9e54bd6d31a | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-9100000000-94daf4b2f125f3a08fc7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0kai-2900000000-821aeeb1835bf094beb8 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0f6t-1900000000-4c7499267ca09c35d1fc | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0udj-1900000000-c8febb206f8338bc0103 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00di-8900000000-ed5cee25f7460f6dee63 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0007-7900000000-d4ba86036408b6707652 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0f6w-5900000000-98925ed00d83d0cb75d2 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-006x-9200000000-a9e17e100c4c54504073 | 2016-08-03 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0uea-0900000000-f1f33c2d1c43e09835ec | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-5900000000-c0032d5daf3b2b7fe72a | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4i-5900000000-92d483635221139105e7 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-f9689d19a8a4fb34dc2c | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-f9689d19a8a4fb34dc2c | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-f9689d19a8a4fb34dc2c | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Selenoamino Acid Metabolism |  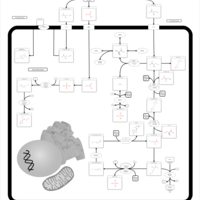  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003966 |
|---|
| DrugBank ID | DB11142 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB012156 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C05335 |
|---|
| BioCyc ID | SELENOMETHIONINE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Selenomethionine |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 105024 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30021 |
|---|
| References |
|---|
| General References | Not Available |
|---|
