| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:44:27 UTC |
|---|
| Updated at | 2020-12-07 19:11:08 UTC |
|---|
| CannabisDB ID | CDB004862 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Homocysteine |
|---|
| Description | Homocysteine, also known as Hcy or homo-cys, belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. A homocysteine that has L configuration. Homocysteine is a very strong basic compound (based on its pKa). Homocysteine exists in all living species, ranging from bacteria to humans. Homocysteine is a potentially toxic compound. Homocysteine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Hcy | ChEBI | | L-2-Amino-4-mercaptobutyric acid | ChEBI | | L-2-Amino-4-mercaptobutyrate | Generator | | 2-Amino-4-mercaptobutyric acid | HMDB | | Homocysteine, L isomer | HMDB | | L-Isomer homocysteine | HMDB | | 2 Amino 4 mercaptobutyric acid | HMDB | | Homocysteine, L-isomer | HMDB | | (S)-2-Amino-4-mercapto-butanoate | HMDB | | (S)-2-Amino-4-mercapto-butanoic acid | HMDB | | (S)-2-Amino-4-mercaptobutanoic acid | HMDB | | (S)-Homocysteine | HMDB | | 2-Amino-4-mercapto-DL-butyrate | HMDB | | 2-Amino-4-mercapto-DL-butyric acid | HMDB | | 2-Amino-4-mercapto-L-butyric acid | HMDB | | 2-Amino-4-mercapto-butanoate | HMDB | | 2-Amino-4-mercapto-butanoic acid | HMDB | | 2-Amino-4-mercapto-butyric acid | HMDB | | 2-Amino-4-sulfanylbutanoate | HMDB | | 2-Amino-4-sulfanylbutanoic acid | HMDB | | D,L-Homocysteine | HMDB | | DL-2-Amino-4-mercapto-butyric acid | HMDB | | DL-2-Amino-4-mercaptobutyric acid | HMDB | | DL-Homocysteine | HMDB | | Homo-cys | HMDB | | L-2-Amino-4-mercapto-butyric acid | HMDB | | L-Homocysteine | HMDB | | 2-Amino-4-mercapto-butyrate | HMDB | | Homocysteine | MeSH, KEGG |
|
|---|
| Chemical Formula | C4H9NO2S |
|---|
| Average Molecular Weight | 135.19 |
|---|
| Monoisotopic Molecular Weight | 135.0354 |
|---|
| IUPAC Name | (2S)-2-amino-4-sulfanylbutanoic acid |
|---|
| Traditional Name | L-homocysteine |
|---|
| CAS Registry Number | 6027-13-0 |
|---|
| SMILES | N[C@@H](CCS)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H9NO2S/c5-3(1-2-8)4(6)7/h3,8H,1-2,5H2,(H,6,7)/t3-/m0/s1 |
|---|
| InChI Key | FFFHZYDWPBMWHY-VKHMYHEASA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Thia fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Alkylthiol
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Hydrocarbon derivative
- Organic oxide
- Primary amine
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Organopnictogen compound
- Primary aliphatic amine
- Organic oxygen compound
- Carbonyl group
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 232 - 233 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 148 mg/mL | Not Available | | logP | -2.56 | Wikipedia |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | Homocysteine, non-derivatized, GC-MS Spectrum | splash10-004i-0920000000-20fbffbe76510c066e6f | Spectrum | | Predicted GC-MS | Homocysteine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-1f800d28bca4b2b7a49f | Spectrum | | Predicted GC-MS | Homocysteine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Homocysteine, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014l-4900000000-c50d79ba3a5013bcf29f | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-000i-9000000000-025c9e1f4fb58138c43b | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT , positive | splash10-014i-4900000000-33f205e002f49f034764 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-052r-0900000000-c1f98c8182b5ddc09cb1 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00ko-9800000000-80a1c80328e8836233a4 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9100000000-1f611da44349ffdfaefe | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0bvi-9000000000-c9d62a66522e46a17f56 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-3900000000-e8322d5858cba16a3bd2 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-00lr-6900000000-e0190a1cc33e605bac0a | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-900bb5d8fb11bec02e39 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-0900000000-38e427e24202b2411597 | 2021-09-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-7900000000-5cf2133d9faac56032d3 | 2021-09-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-942ac689538269d6ca7b | 2021-09-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9000000000-166428542631f9ac0714 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-9000000000-9bcb57b924579a6c3411 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4j-9000000000-8eee886864db64f3eec2 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Methionine Metabolism | 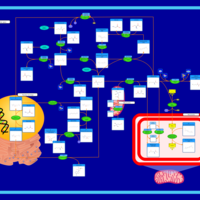 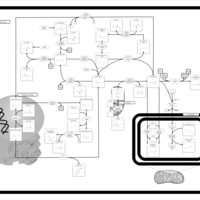 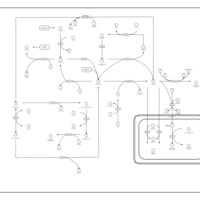 |  | | Glycine and Serine Metabolism | 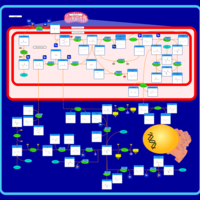 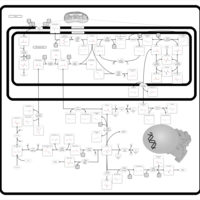 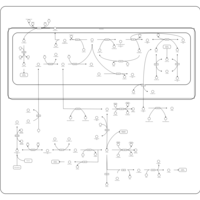 |  | | Betaine Metabolism |  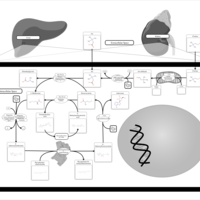  |  | | Homocysteine Degradation | 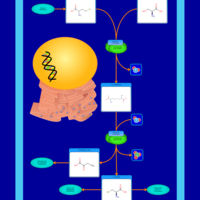 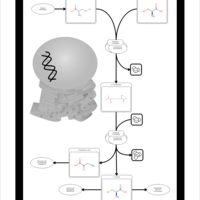 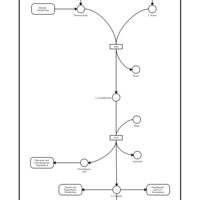 | Not Available | | Catecholamine Biosynthesis | 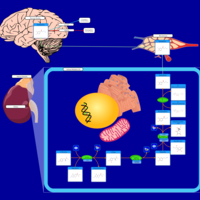 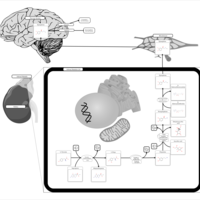  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000742 |
|---|
| DrugBank ID | DB04422 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001491 |
|---|
| KNApSAcK ID | C00001365 |
|---|
| Chemspider ID | 82666 |
|---|
| KEGG Compound ID | C00155 |
|---|
| BioCyc ID | HOMO-CYS |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Homocysteine |
|---|
| METLIN ID | 3256 |
|---|
| PubChem Compound | 91552 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17588 |
|---|
| References |
|---|
| General References | Not Available |
|---|


