| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:39:24 UTC |
|---|
| Updated at | 2022-12-13 23:36:23 UTC |
|---|
| CannabisDB ID | CDB004813 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Asparagine |
|---|
| Description | L-Asparagine, also known as Asn or aspartamic acid, belongs to the class of organic compounds known as asparagine and derivatives. Asparagine and derivatives are compounds containing asparagine or a derivative thereof resulting from reaction of asparagine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. L-Asparagine is a drug which is used for nutritional supplementation, also for treating dietary shortage or imbalance. L-Asparagine is a very strong basic compound (based on its pKa). L-Asparagine exists in all living species, ranging from bacteria to humans. Within humans, L-asparagine participates in a number of enzymatic reactions. In particular, L-asparagine and L-glutamic acid can be biosynthesized from L-aspartic acid and L-glutamine through the action of the enzyme asparagine synthetase [glutamine-hydrolyzing]. In addition, L-asparagine can be converted into L-aspartic acid through the action of the enzyme isoaspartyl peptidase/l-asparaginase. In humans, L-asparagine is involved in aspartate metabolism. L-Asparagine is an odorless tasting compound. Outside of the human body, L-Asparagine is found, on average, in the highest concentration within a few different foods, such as white lupines, wheats, and oats and in a lower concentration in sacred lotus, parsnips, and pineapples. L-Asparagine has also been detected, but not quantified in, several different foods, such as colorado pinyons, opium poppies, lentils, green beans, and watermelons. This could make L-asparagine a potential biomarker for the consumption of these foods. L-Asparagine is a potentially toxic compound. An optically active form of asparagine having L-configuration. L-Asparagine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (2S)-2,4-Diamino-4-oxobutanoic acid | ChEBI | | (2S)-2-Amino-3-carbamoylpropanoic acid | ChEBI | | (S)-2-Amino-3-carbamoylpropanoic acid | ChEBI | | (S)-Asparagine | ChEBI | | 2-Aminosuccinamic acid | ChEBI | | alpha-Aminosuccinamic acid | ChEBI | | Asn | ChEBI | | ASPARAGINE | ChEBI | | Aspartamic acid | ChEBI | | L-2-Aminosuccinamic acid | ChEBI | | L-Asparagin | ChEBI | | L-Aspartic acid beta-amide | ChEBI | | N | ChEBI | | (2S)-2,4-Diamino-4-oxobutanoate | Generator | | (2S)-2-Amino-3-carbamoylpropanoate | Generator | | (S)-2-Amino-3-carbamoylpropanoate | Generator | | 2-Aminosuccinamate | Generator | | a-Aminosuccinamate | Generator | | a-Aminosuccinamic acid | Generator | | alpha-Aminosuccinamate | Generator | | Α-aminosuccinamate | Generator | | Α-aminosuccinamic acid | Generator | | Aspartamate | Generator | | L-2-Aminosuccinamate | Generator | | L-Aspartate b-amide | Generator | | L-Aspartate beta-amide | Generator | | L-Aspartate β-amide | Generator | | L-Aspartic acid b-amide | Generator | | L-Aspartic acid β-amide | Generator | | (-)-Asparagine | HMDB | | (S)-2,4-Diamino-4-oxobutanoate | HMDB | | (S)-2,4-Diamino-4-oxobutanoic acid | HMDB | | Agedoite | HMDB | | alpha Amminosuccinamate | HMDB | | alpha Amminosuccinamic acid | HMDB | | Altheine | HMDB | | Asparagine acid | HMDB | | Asparamide | HMDB | | Aspartic acid amide | HMDB | | Aspartic acid b-amide | HMDB | | Aspartic acid beta amide | HMDB | | b2,4-(S)-Diamino-4-oxo-utanoate | HMDB | | b2,4-(S)-Diamino-4-oxo-utanoic acid | HMDB | | Crystal VI | HMDB | | L-2,4-Diamino-4-oxobutanoate | HMDB | | L-2,4-Diamino-4-oxobutanoic acid | HMDB | | L-Aspartamine | HMDB | | L-b-Asparagine | HMDB | | L-beta-Asparagine | HMDB |
|
|---|
| Chemical Formula | C4H8N2O3 |
|---|
| Average Molecular Weight | 132.12 |
|---|
| Monoisotopic Molecular Weight | 132.0535 |
|---|
| IUPAC Name | (2S)-2-amino-3-carbamoylpropanoic acid |
|---|
| Traditional Name | L-asparagine |
|---|
| CAS Registry Number | 70-47-3 |
|---|
| SMILES | N[C@@H](CC(N)=O)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C4H8N2O3/c5-2(4(8)9)1-3(6)7/h2H,1,5H2,(H2,6,7)(H,8,9)/t2-/m0/s1 |
|---|
| InChI Key | DCXYFEDJOCDNAF-REOHCLBHSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as asparagine and derivatives. Asparagine and derivatives are compounds containing asparagine or a derivative thereof resulting from reaction of asparagine at the amino group or the carboxy group, or from the replacement of any hydrogen of glycine by a heteroatom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | Asparagine and derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Asparagine or derivatives
- Alpha-amino acid
- L-alpha-amino acid
- Fatty amide
- Fatty acyl
- Fatty acid
- Carboxamide group
- Amino acid
- Primary carboxylic acid amide
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic nitrogen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Carbonyl group
- Amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Environmental role: Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 234 - 235 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 29.4 mg/mL | Not Available | | logP | -3.82 | CHMELIK,J ET AL. (1991) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Asparagine, 3 TMS, GC-MS Spectrum | splash10-0159-0910000000-bedf57998656ab5ebc16 | Spectrum | | GC-MS | L-Asparagine, 3 TMS, GC-MS Spectrum | splash10-0159-0910000000-4979c4d028d2dc931263 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-b7ea3fef61f3940cbdda | Spectrum | | GC-MS | L-Asparagine, 3 TMS, GC-MS Spectrum | splash10-00di-9710000000-6d0afcbcc003347e598e | Spectrum | | GC-MS | L-Asparagine, 2 TMS, GC-MS Spectrum | splash10-0uxr-0910000000-2be567239bd3229b1ca1 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-02t9-0790000000-b8141f48cbebb90f683e | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-02t9-1790000000-1d12ed9b4fb2799da766 | Spectrum | | GC-MS | L-Asparagine, 4 TMS, GC-MS Spectrum | splash10-014u-0961000000-65a5c0999f17110c9939 | Spectrum | | GC-MS | L-Asparagine, 4 TMS, GC-MS Spectrum | splash10-00kr-0940000000-0e0e5c7bdbac5ea0e49e | Spectrum | | GC-MS | L-Asparagine, 3 TMS, GC-MS Spectrum | splash10-00lr-1920000000-7864dbb1f685e64dd1cf | Spectrum | | GC-MS | L-Asparagine, 4 TMS, GC-MS Spectrum | splash10-000i-1940000000-964ea25da2789805985a | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-00lr-0930000000-79a089d9875e092dd7f2 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-bedf57998656ab5ebc16 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-4979c4d028d2dc931263 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-0159-0910000000-b7ea3fef61f3940cbdda | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-00di-4931100000-17f149310ce3a0f748d1 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-00di-9710000000-6d0afcbcc003347e598e | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-0uxr-0910000000-2be567239bd3229b1ca1 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-02t9-0790000000-b8141f48cbebb90f683e | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-02t9-1790000000-1d12ed9b4fb2799da766 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-014u-0961000000-65a5c0999f17110c9939 | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-00kr-0940000000-0e0e5c7bdbac5ea0e49e | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-00lr-1920000000-7864dbb1f685e64dd1cf | Spectrum | | GC-MS | L-Asparagine, non-derivatized, GC-MS Spectrum | splash10-000i-1940000000-964ea25da2789805985a | Spectrum | | Predicted GC-MS | L-Asparagine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-000f-9100000000-5881591331bd059b7e7d | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0080-9400000000-e5c7e19f427eea6d71b0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00di-9000000000-afc1214100db1168b095 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00dl-9000000000-57e977cd87e9a86d482b | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6fb96f5aa291359dba29 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-76f11e6fe5657c35d15f | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-e944486273dbfde4cae4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-27ad91a86be0c4d86c3c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-b8b7a3431b66246ad613 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-03fa294ec740e189dd99 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-000i-9000000000-cee2081406fc25ab6169 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-cd79afa0c27f65e54adc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-01q9-0943200000-5a24322ba0ce2f410155 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0900000000-fc3086cc1bb0c06a4c11 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-3900000000-2d74d8232e7523f3b1c9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-fa61c6fa9a87ce4905bb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-01q9-0942200000-757da8b3406485c997b0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-014i-9000000000-eac18512f2d22ab7a7ef | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0900000000-8df6d5b6cf94bf081c89 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-001i-0900000000-18079004a95c252da208 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-001i-0900000000-e6b1f9b4982e6cdc863d | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-03di-5900000000-7b9c09b3b6de28972f97 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00dl-9100000000-2ea95ead344d62e230a4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-defacc365589bc4437d7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-fd60db4d5e35794c45ae | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-001i-3900000000-fd0f1034b5c1e40435c0 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Aspartate Metabolism |    |  | | Ammonia Recycling |   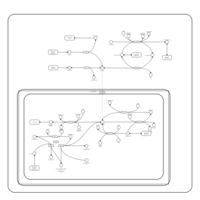 |  | | Canavan Disease | 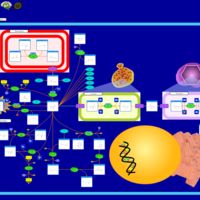 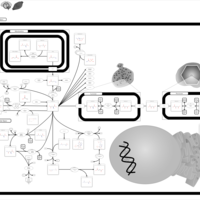 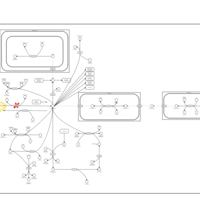 | Not Available | | Hypoacetylaspartia |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Asparagine synthetase [glutamine-hydrolyzing] | ASNS | 7q21.3 | P08243 | details | | Asparagine--tRNA ligase, cytoplasmic | NARS | 18q21.31 | O43776 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1 | RPN1 | 3q21.3 | P04843 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit | DDOST | 1p36.1 | P39656 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 2 | RPN2 | 20q12-q13.1 | P04844 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A | STT3A | 11q23.3 | P46977 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1 | DAD1 | 14q11.2 | P61803 | details | | Isoaspartyl peptidase/L-asparaginase | ASRGL1 | 11q12.3 | Q7L266 | details | | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B | STT3B | 3p23 | Q8TCJ2 | details | | 60 kDa lysophospholipase | ASPG | 14q32.33 | Q86U10 | details | | Probable asparagine--tRNA ligase, mitochondrial | NARS2 | 11q14.1 | Q96I59 | details |
|
|---|
| Transporters | |
| Neutral amino acid transporter B(0) | SLC1A5 | 19q13.3 | Q15758 | details | | Sodium-coupled neutral amino acid transporter 3 | SLC38A3 | 3p21.3 | Q99624 | details |
|
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| |
| Alien Dawg | Detected and Quantified | 36.333 mg/g dry wt | | details | | Gabriola | Detected and Quantified | 16.370 mg/g dry wt | | details | | Island Honey | Detected and Quantified | 16.925 mg/g dry wt | | details | | Quadra | Detected and Quantified | 13.542 mg/g dry wt | | details | | Sensi Star | Detected and Quantified | 27.005 mg/g dry wt | | details | | Tangerine Dream | Detected and Quantified | 13.159 mg/g dry wt | | details |
|
|---|
| External Links |
|---|
| HMDB ID | HMDB0000168 |
|---|
| DrugBank ID | DB00174 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000787 |
|---|
| KNApSAcK ID | C00001341 |
|---|
| Chemspider ID | 6031 |
|---|
| KEGG Compound ID | C00152 |
|---|
| BioCyc ID | ASN |
|---|
| BiGG ID | 34055 |
|---|
| Wikipedia Link | Asparagine |
|---|
| METLIN ID | 14 |
|---|
| PubChem Compound | 6267 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17196 |
|---|
| References |
|---|
| General References | Not Available |
|---|

