| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:25:22 UTC |
|---|
| Updated at | 2020-11-18 16:39:37 UTC |
|---|
| CannabisDB ID | CDB005260 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | D-Fructose 2,6-bisphosphate |
|---|
| Description | D-Fructose 2,6-bisphosphate, also known as phosphofructokinase activator, belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. A D-fructofuranose 2,6-bisphosphate with a beta-configuration at the anomeric centre. D-Fructose 2,6-bisphosphate is an extremely weak basic (essentially neutral) compound (based on its pKa). D-Fructose 2,6-bisphosphate exists in all living species, ranging from bacteria to humans. D-fructose 2,6-bisphosphate can be converted into fructose 6-phosphate; which is mediated by the enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1. In humans, D-fructose 2,6-bisphosphate is involved in the metabolic disorder called fructosuria. D-Fructose 2,6-bisphosphate is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,6-Di-O-phosphono-beta-D-fructofuranose | ChEBI | | 2,6-Di-O-phosphono-b-D-fructofuranose | Generator | | 2,6-Di-O-phosphono-β-D-fructofuranose | Generator | | D-Fructose 2,6-bisphosphoric acid | Generator | | b-D-Fructose 2,6-bisphosphate | HMDB | | beta-D-Fructose 2,6-bisphosphate | HMDB | | Fru 2,6-P2, fructose 2,6-diphosphate | HMDB | | Fructose 2,6-bisphosphate | HMDB | | Fructose 2,6-biphosphate | MeSH, HMDB | | Fructose 2,6-diphosphate | MeSH, HMDB | | Phosphofructokinase activation factor | MeSH, HMDB | | Phosphofructokinase activator | MeSH, HMDB | | Fructose-2,6-diphosphate | MeSH, HMDB | | D-Fructose 2,6-diphosphate | HMDB | | beta-D-Fructose 2,6-diphosphate | HMDB | | β-D-Fructose 2,6-diphosphate | HMDB |
|
|---|
| Chemical Formula | C6H14O12P2 |
|---|
| Average Molecular Weight | 340.12 |
|---|
| Monoisotopic Molecular Weight | 339.996 |
|---|
| IUPAC Name | {[(2R,3S,4S,5S)-3,4-dihydroxy-5-(hydroxymethyl)-5-(phosphonooxy)oxolan-2-yl]methoxy}phosphonic acid |
|---|
| Traditional Name | fructose-2,6-diphosphate |
|---|
| CAS Registry Number | Not Available |
|---|
| SMILES | OC[C@@]1(OP(O)(O)=O)O[C@H](COP(O)(O)=O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H14O12P2/c7-2-6(18-20(13,14)15)5(9)4(8)3(17-6)1-16-19(10,11)12/h3-5,7-9H,1-2H2,(H2,10,11,12)(H2,13,14,15)/t3-,4-,5+,6+/m1/s1 |
|---|
| InChI Key | YXWOAJXNVLXPMU-ZXXMMSQZSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pentose phosphates. These are carbohydrate derivatives containing a pentose substituted by one or more phosphate groups. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Pentose phosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pentose phosphate
- Pentose-5-phosphate
- C-glycosyl compound
- Glycosyl compound
- Monosaccharide phosphate
- Monoalkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Alkyl phosphate
- Tetrahydrofuran
- Secondary alcohol
- Organoheterocyclic compound
- Oxacycle
- Primary alcohol
- Alcohol
- Hydrocarbon derivative
- Organic oxide
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, GC-MS Spectrum | splash10-01dj-0963000000-090736f476a618727455 | Spectrum | | GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, GC-MS Spectrum | splash10-00tb-0972000000-e47c74a14376eaabd8c3 | Spectrum | | GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, GC-MS Spectrum | splash10-01dj-0963000000-090736f476a618727455 | Spectrum | | GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, GC-MS Spectrum | splash10-00tb-0972000000-e47c74a14376eaabd8c3 | Spectrum | | Predicted GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0002-9512000000-82418e0e398490357c5e | Spectrum | | Predicted GC-MS | D-Fructose 2,6-bisphosphate, 3 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-2190010000-53cf738181713979e181 | Spectrum | | Predicted GC-MS | D-Fructose 2,6-bisphosphate, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ 10V, negative | splash10-000i-0019000000-7b58b1d6169f22e732a8 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ 20V, negative | splash10-000m-2496000000-40ad2c4944ae5c0701f3 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ 30V, negative | splash10-0002-9830000000-a9cbb8a3dba391a8858b | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ 40V, negative | splash10-002b-9200000000-6dcaff4e5eee3799656f | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ 50V, negative | splash10-004j-9100000000-9851b59e56acb7c5802c | 2020-07-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-3797000000-cb11e27ce8d0be902a39 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-9132000000-b908246df46f54e31180 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00dm-9600000000-19262e5981934144e1aa | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-7409000000-0240bd8771cde252759c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9000000000-8930f98e412f1b68e30b | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-fb204edc71835bcbaaad | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0393000000-f7f716e45f745f0bec68 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0002-1920000000-f0eccbb835449758a67c | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0f6t-9410000000-f9f76bd919e86701aaff | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002r-9008000000-d519306c146482d051d0 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004i-9001000000-2d0e9375a49babbe9d93 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-bc05f33d90e2bf18235c | 2021-09-24 | View Spectrum |
|
|---|
| NMR | Not Available |
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Fructose and Mannose Degradation | 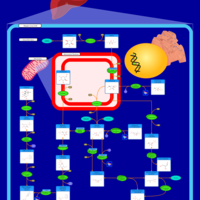 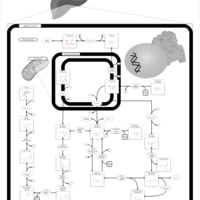 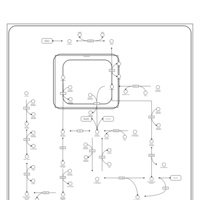 |  | | Fructosuria | 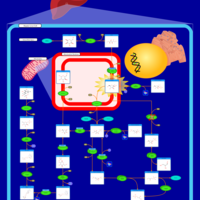   | Not Available | | Fructose intolerance, hereditary |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 | PFKFB3 | 10p15.1 | Q16875 | details | | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 | PFKFB4 | 3p22-p21 | Q16877 | details | | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 1 | PFKFB1 | Xp11.21 | P16118 | details | | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 | PFKFB2 | 1q31 | O60825 | details | | Fructose-2,6-bisphosphatase TIGAR | TIGAR | 12p13.3 | Q9NQ88 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001047 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022390 |
|---|
| KNApSAcK ID | C00007441 |
|---|
| Chemspider ID | 94762 |
|---|
| KEGG Compound ID | C00665 |
|---|
| BioCyc ID | CPD-535 |
|---|
| BiGG ID | 35645 |
|---|
| Wikipedia Link | Fructose_2,6-bisphosphate |
|---|
| METLIN ID | 5964 |
|---|
| PubChem Compound | 105021 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 28602 |
|---|
| References |
|---|
| General References | Not Available |
|---|
