| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:22:52 UTC |
|---|
| Updated at | 2020-11-18 16:39:35 UTC |
|---|
| CannabisDB ID | CDB005235 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | D-Galactose |
|---|
| Description | D-galactose, also known as galactopyranose, is an aldohexose is a C4-epimer of glucose. It is as sweet as glucose and only 65% as sweet as sucrose. When D-galactose condenses with glucose, lactose if formed. In humans and other species, galactose is metabolized to glucose largely by the Leloir pathway. D-galactose is converted to UDP-glucose and glucose-1-phosphate by three enzymes (PMID: 1292318 ):  1) Galactokinase (GALK; KEGG enzyme E.C. 2.7.1.6) phosphorylates D-galactose to D-galactose-1-phosphate; 2) Then D-galactose-1-phosphate uridyltransferase (GALT; KEGG enzyme E.C. 2.7.7.12) transfers a UMP from UDP-glucose to D-galactose-1-phosphate to form UDP-galactose and glucose-1-phosphate. Glucose-1-phosphate will then enter the glycolytic pathway for energy production. 3) UDP-galactose epimerase (GALE) then interconverts UDP-galactose to UDP-glucose. Lactose in mammalian milk is the main source of D-galactose. Galactose in the urine is a biomarker for the consumption of milk. Galactose can also be found in some fruits and vegetables such as in banana, orange, papaya, bell pepper, avocado and sugar beets ( Ref:DOI ). It is also found in fig leaves, sesame, carob bean, caraway seeds and beet, cocoa, chive, potato, and cannabis plants (PMID: 6991645 ). In addition to D-Galactose providing energy for the body, it is also a necessary substrate for the biosynthesis of many macromolecules such as cerebrosides, gangliosides, and mucoproteins. Galactose is a component of the antigens of the ABO blood group (PMID: 15373665 ). Metabolic pathways for D-galactose are important not only for the provision of these pathways but also for the prevention of D-galactose accumulation. There are diseases resulting from the deficiencies of the three enzymes of the Leloir pathway. Classic galactosemia, or the accumulation of D-galactose in the blood, is the rare inborn error of D-galactose metabolism, caused by the deficiency of the second enzyme GALT, (from mutations in the GALT gene; PMID: 10408771 ). As galactose accumulation is associated with health problems (PMID: 26001656 ), particularly involving the liver (PMID: 29502917 ), the discovery of alternative metabolic pathways to the Leloir in fungi and bacteria (PMID:15256214 ) could lead to discovery of analogous genes in humans. D-galactose was also be protective against pancreatitis (PMID: 2989374 ) and streptozotocin-induced model of Alzheimer’s disease in rats (PMID: 29501615 ). D-Galactose is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| ALPHA D-GALACTOSE | ChEBI | | alpha-D-Gal | ChEBI | | Gal-alpha | ChEBI | | alpha-D-Galactose | Kegg | | a D-GALACTOSE | Generator | | Α D-galactose | Generator | | a-D-Gal | Generator | | Α-D-gal | Generator | | Gal-a | Generator | | Gal-α | Generator | | a-D-Galactose | Generator | | Α-D-galactose | Generator | | (+)-Galactose | HMDB | | 5Abp | HMDB | | 8Abp | HMDB | | alpha-D-Galactopyranose | HMDB | | D-(+)-Galactose | HMDB | | D-Hexose | HMDB | | GAL | HMDB | | Galactose | HMDB | | Galactose (NF) | HMDB | | GLA | HMDB | | GLC | HMDB | | Hexose | HMDB | | SHU 508 a | MeSH, HMDB | | SH-TA-508 | MeSH, HMDB | | SHU 508 | MeSH, HMDB | | Levovist | MeSH, HMDB | | SHU-508 | MeSH, HMDB |
|
|---|
| Chemical Formula | C6H12O6 |
|---|
| Average Molecular Weight | 180.16 |
|---|
| Monoisotopic Molecular Weight | 180.0634 |
|---|
| IUPAC Name | (2S,3R,4S,5R,6R)-6-(hydroxymethyl)oxane-2,3,4,5-tetrol |
|---|
| Traditional Name | galactose |
|---|
| CAS Registry Number | 3646-73-9 |
|---|
| SMILES | OC[C@H]1O[C@H](O)[C@H](O)[C@@H](O)[C@H]1O |
|---|
| InChI Identifier | InChI=1S/C6H12O6/c7-1-2-3(8)4(9)5(10)6(11)12-2/h2-11H,1H2/t2-,3+,4+,5-,6+/m1/s1 |
|---|
| InChI Key | WQZGKKKJIJFFOK-PHYPRBDBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as hexoses. These are monosaccharides in which the sugar unit is a is a six-carbon containing moeity. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | Hexoses |
|---|
| Alternative Parents | |
|---|
| Substituents | - Hexose monosaccharide
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 170 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 683 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-066s-0941000000-8f1b68655e0a5ed179e3 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ldj-0941000000-84c771fab416aa0a40d8 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ktb-0931000000-6f37a774d1ab64585c8d | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-066s-0931000000-9945093085ef7b0de618 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ldj-0931000000-1ed532d5d2897be775df | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-00di-9731000000-af5bd67ad0bbb2997839 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-00di-9621000000-9dad5bd1760a891ace2a | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0fr2-0920000000-d0bbaf502997c43971ba | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0fr2-1920000000-f47f0b66263744db15ba | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-066s-0941000000-8f1b68655e0a5ed179e3 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ldj-0941000000-84c771fab416aa0a40d8 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ktb-0931000000-6f37a774d1ab64585c8d | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-066s-0931000000-9945093085ef7b0de618 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0ldj-0931000000-1ed532d5d2897be775df | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-00di-9731000000-af5bd67ad0bbb2997839 | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-00di-9621000000-9dad5bd1760a891ace2a | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0fr2-0920000000-d0bbaf502997c43971ba | Spectrum | | GC-MS | D-Galactose, non-derivatized, GC-MS Spectrum | splash10-0fr2-1920000000-f47f0b66263744db15ba | Spectrum | | Predicted GC-MS | D-Galactose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0np0-9700000000-e8d638dc817e46b97d7b | Spectrum | | Predicted GC-MS | D-Galactose, 5 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-004i-6122690000-eaf6f7adf34ccd0c667b | Spectrum | | Predicted GC-MS | D-Galactose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Galactose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Galactose, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Galactose, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | D-Galactose, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0f6t-0900000000-70d3989fddd34b894c77 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00ri-9700000000-d454170b4e3ed18ed2d9 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-007a-9300000000-81f5f12f84924a34e2c0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 10V, negative | splash10-059i-9100000000-33aacf4c62b17cfa394e | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 20V, negative | splash10-0a4i-9000000000-79ea696263aaad168701 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF 40V, negative | splash10-0a4i-9000000000-480461c1cdd9d138dce2 | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-059i-9100000000-33aacf4c62b17cfa394e | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-9000000000-79ea696263aaad168701 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-9000000000-480461c1cdd9d138dce2 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01q9-0900000000-b0bc47623e7b2ca31c02 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03ea-3900000000-648e1637af29cf2a3518 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0007-9200000000-9e6f46a1cbf52d6e347a | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-a4ec4f0b1e29e360a952 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01t9-6900000000-7b3ea9c64ecc8d4ac867 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-052f-9100000000-ec2bf4918640a0a36398 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-2900000000-2448926b508622464fe7 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-056r-8900000000-4c073cb93b78120113e6 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-ece70093ab5d3c331ac4 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-01qa-0900000000-04ceb34d441ff6a75763 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03dl-9400000000-a8f1ceab155611f949c1 | 2021-09-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ow-9000000000-358f68fc2b7a72c27546 | 2021-09-25 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 400 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Galactose Metabolism | 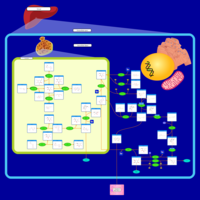 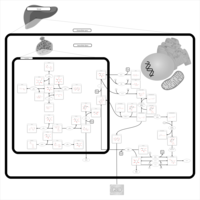 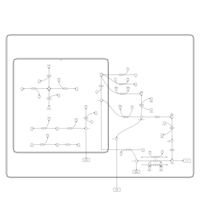 |  | | Nucleotide Sugars Metabolism | 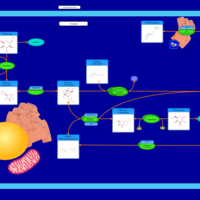 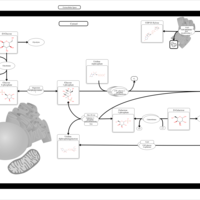 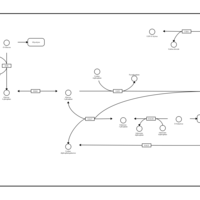 |  | | Lactose Degradation | 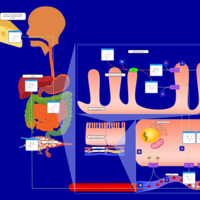 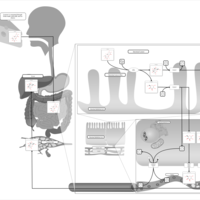 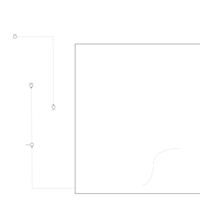 | Not Available | | Galactosemia II (GALK) | 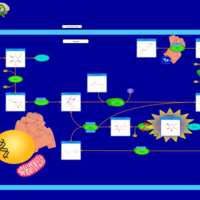 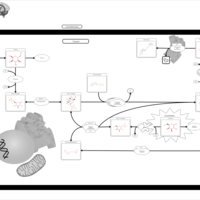 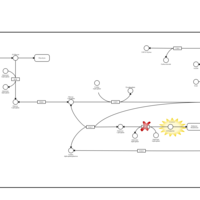 | Not Available | | Galactosemia III | 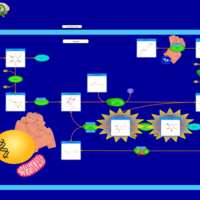 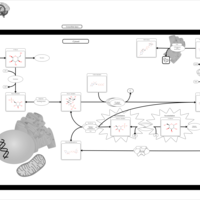 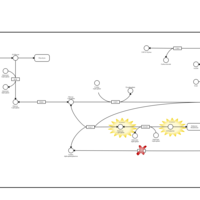 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000143 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB021787 |
|---|
| KNApSAcK ID | C00001119 |
|---|
| Chemspider ID | Not Available |
|---|
| KEGG Compound ID | C00984 |
|---|
| BioCyc ID | ALPHA-D-GALACTOSE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Galactose |
|---|
| METLIN ID | Not Available |
|---|
| PubChem Compound | 439357 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 28061 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
- Tyfield L, Reichardt J, Fridovich-Keil J, Croke DT, Elsas LJ 2nd, Strobl W, Kozak L, Coskun T, Novelli G, Okano Y, Zekanowski C, Shin Y, Boleda MD: Classical galactosemia and mutations at the galactose-1-phosphate uridyl transferase (GALT) gene. Hum Mutat. 1999;13(6):417-30. doi: 10.1002/(SICI)1098-1004(1999)13:6<417::AID-HUMU1>3.0.CO;2-0. [PubMed:10408771 ]
- Lai K, Klapa MI: Alternative pathways of galactose assimilation: could inverse metabolic engineering provide an alternative to galactosemic patients? Metab Eng. 2004 Jul;6(3):239-44. doi: 10.1016/j.ymben.2004.01.001. [PubMed:15256214 ]
- Blumenreich GA: The standard of care. AANA J. 1992 Dec;60(6):529-31. [PubMed:1292318 ]
- Yamamoto F: Review: ABO blood group system--ABH oligosaccharide antigens, anti-A and anti-B, A and B glycosyltransferases, and ABO genes. Immunohematology. 2004;20(1):3-22. [PubMed:15373665 ]
- Coelho AI, Berry GT, Rubio-Gozalbo ME: Galactose metabolism and health. Curr Opin Clin Nutr Metab Care. 2015 Jul;18(4):422-7. doi: 10.1097/MCO.0000000000000189. [PubMed:26001656 ]
- Demirbas D, Brucker WJ, Berry GT: Inborn Errors of Metabolism with Hepatopathy: Metabolism Defects of Galactose, Fructose, and Tyrosine. Pediatr Clin North Am. 2018 Apr;65(2):337-352. doi: 10.1016/j.pcl.2017.11.008. [PubMed:29502917 ]
- Van Meel FC, Steenbakkers PG, Oomen JC: Human and chimpanzee monoclonal antibodies. J Immunol Methods. 1985 Jun 25;80(2):267-76. doi: 10.1016/0022-1759(85)90027-4. [PubMed:2989374 ]
- Knezovic A, Osmanovic Barilar J, Babic A, Bagaric R, Farkas V, Riederer P, Salkovic-Petrisic M: Glucagon-like peptide-1 mediates effects of oral galactose in streptozotocin-induced rat model of sporadic Alzheimer's disease. Neuropharmacology. 2018 Jun;135:48-62. doi: 10.1016/j.neuropharm.2018.02.027. Epub 2018 Feb 28. [PubMed:29501615 ]
|
|---|

