| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:12:51 UTC |
|---|
| Updated at | 2020-12-07 19:11:38 UTC |
|---|
| CannabisDB ID | CDB005135 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Glutaryl-CoA |
|---|
| Description | Glutaryl-CoA belongs to the class of organic compounds known as long-chain fatty acyl coas. These are acyl CoAs where the group acylated to the coenzyme A moiety is a long aliphatic chain of 13 to 21 carbon atoms. Glutaryl-CoA is a very strong basic compound (based on its pKa). Glutaryl-CoA is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Glutaryl-coenzyme A | HMDB |
|
|---|
| Chemical Formula | C26H42N7O19P3S |
|---|
| Average Molecular Weight | 881.63 |
|---|
| Monoisotopic Molecular Weight | 881.1469 |
|---|
| IUPAC Name | 5-{[2-(3-{3-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-2-hydroxy-3-methylbutanamido}propanamido)ethyl]sulfanyl}-5-oxopentanoic acid |
|---|
| Traditional Name | 5-({2-[3-(3-{[({[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methoxy(hydroxy)phosphoryl}oxy(hydroxy)phosphoryl)oxy]methyl}-2-hydroxy-3-methylbutanamido)propanamido]ethyl}sulfanyl)-5-oxopentanoic acid |
|---|
| CAS Registry Number | 103192-48-9 |
|---|
| SMILES | CC(C)(COP(O)(=O)OP(O)(=O)OC[C@H]1O[C@H]([C@H](O)[C@@H]1OP(O)(O)=O)N1C=NC2=C1N=CN=C2N)C(O)C(=O)NCCC(=O)NCCSC(=O)CCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C26H42N7O19P3S/c1-26(2,21(39)24(40)29-7-6-15(34)28-8-9-56-17(37)5-3-4-16(35)36)11-49-55(46,47)52-54(44,45)48-10-14-20(51-53(41,42)43)19(38)25(50-14)33-13-32-18-22(27)30-12-31-23(18)33/h12-14,19-21,25,38-39H,3-11H2,1-2H3,(H,28,34)(H,29,40)(H,35,36)(H,44,45)(H,46,47)(H2,27,30,31)(H2,41,42,43)/t14-,19-,20-,21?,25-/m1/s1 |
|---|
| InChI Key | SYKWLIJQEHRDNH-KRPIADGTSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as long-chain fatty acyl coas. These are acyl CoAs where the group acylated to the coenzyme A moiety is a long aliphatic chain of 13 to 21 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acyl thioesters |
|---|
| Direct Parent | Long-chain fatty acyl CoAs |
|---|
| Alternative Parents | |
|---|
| Substituents | - Coenzyme a or derivatives
- Purine ribonucleoside 3',5'-bisphosphate
- Purine ribonucleoside bisphosphate
- Purine ribonucleoside diphosphate
- Ribonucleoside 3'-phosphate
- Pentose phosphate
- Pentose-5-phosphate
- Beta amino acid or derivatives
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Monosaccharide phosphate
- Organic pyrophosphate
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Monoalkyl phosphate
- Aminopyrimidine
- Imidolactam
- N-acyl-amine
- N-substituted imidazole
- Organic phosphoric acid derivative
- Monosaccharide
- Pyrimidine
- Alkyl phosphate
- Fatty amide
- Phosphoric acid ester
- Tetrahydrofuran
- Imidazole
- Azole
- Heteroaromatic compound
- Carbothioic s-ester
- Secondary alcohol
- Thiocarboxylic acid ester
- Carboxamide group
- Secondary carboxylic acid amide
- Amino acid or derivatives
- Sulfenyl compound
- Thiocarboxylic acid or derivatives
- Organoheterocyclic compound
- Azacycle
- Oxacycle
- Carboxylic acid derivative
- Organosulfur compound
- Organic oxygen compound
- Hydrocarbon derivative
- Carbonyl group
- Organic nitrogen compound
- Primary amine
- Organopnictogen compound
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Alcohol
- Amine
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | |
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-00kr-0005009000-eb3a971b38526182c8ac | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-0a4i-0269300000-b433e45be2e4ddb9b3dd | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-05n1-0009700000-d04f275432ce90be4c03 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-0a4l-0691000000-1c9c9dcd3392e14e12d9 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-0f80-0009841000-c64bfcd1084f04509fc5 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-000i-0009000000-8ef4f57ae35ca24154ac | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-014r-0001920000-f105e4c7bcf62b49d748 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 61V, negative | splash10-001i-0000100090-7e0c0d1ae140f9f6c57c | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 77V, negative | splash10-053r-0000920060-2e8ff86ba386000c087e | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 100V, negative | splash10-0a6r-1211920000-b87d792a259c80a7b01d | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 123V, negative | splash10-056r-9835710000-f9df91f65d08757ed109 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 146V, negative | splash10-004i-9512100000-6725b9c211e210ffa4f9 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 169V, negative | splash10-004i-9400000000-3b7b4c8aa8db05827423 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 200V, negative | splash10-004i-9300000000-a481ad1113e5856425ad | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 232V, negative | splash10-004i-9200000000-484261819d8fe28a06bc | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Orbitrap 278V, negative | splash10-004i-9100000000-1ced5b391be9b0f5176c | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-001i-0001960370-e5f5990aef17b3100ae7 | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-00di-0190000000-0bdc30775338012cc42f | 2020-07-22 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - n/a 61V, negative | splash10-0a4i-0900000000-816bc86479bd970ceed6 | 2020-07-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-1912000130-9b9c96863df04861fbee | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0913000000-43f2e61c32509ed14b97 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-1901000000-4ff7fbc1bd490ad4c020 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-01q9-3911030350-425e74ceb7a0a4256402 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01si-2901010010-f93605fcb2512efdd305 | 2015-09-15 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-057i-6900100000-74a35eb8b1b0810315e4 | 2015-09-15 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Lysine Degradation | 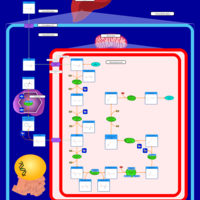 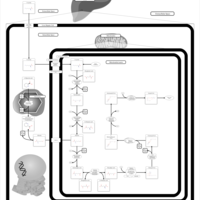 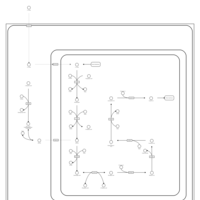 |  | | Fatty acid Metabolism | 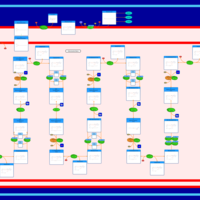  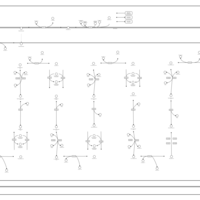 |  | | Ethylmalonic Encephalopathy | 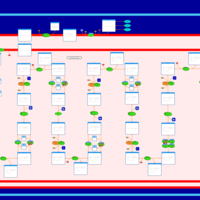  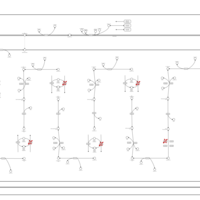 | Not Available | | Glutaric Aciduria Type I | 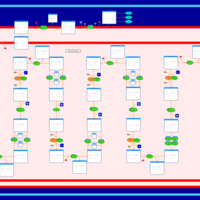  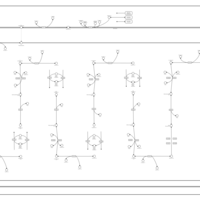 | Not Available | | Short Chain Acyl CoA Dehydrogenase Deficiency (SCAD Deficiency) | 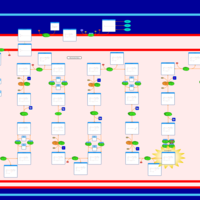 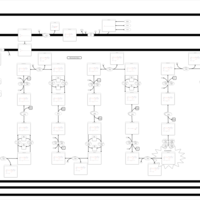 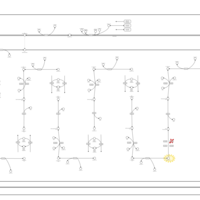 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Dihydrolipoyl dehydrogenase, mitochondrial | DLD | 7q31-q32 | P09622 | details | | Glutaryl-CoA dehydrogenase, mitochondrial | GCDH | 19p13.2 | Q92947 | details | | 2-oxoglutarate dehydrogenase, mitochondrial | OGDH | 7p14-p13 | Q02218 | details | | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | DLST | 14q24.3 | P36957 | details | | 2-oxoglutarate dehydrogenase-like, mitochondrial | OGDHL | | Q9ULD0 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001339 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB022563 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 388388 |
|---|
| KEGG Compound ID | C00527 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | 35255 |
|---|
| Wikipedia Link | Glutaryl-CoA |
|---|
| METLIN ID | 6173 |
|---|
| PubChem Compound | 439252 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 15524 |
|---|
| References |
|---|
| General References | Not Available |
|---|

