| Identification |
|---|
| HMDB Protein ID
| CDBP00052 |
| Secondary Accession Numbers
| Not Available |
| Name
| Dihydrolipoyl dehydrogenase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- Dihydrolipoamide dehydrogenase
- Glycine cleavage system L protein
|
| Gene Name
| DLD |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxidoreductase activity |
| Specific Function
| Lipoamide dehydrogenase is a component of the glycine cleavage system as well as of the alpha-ketoacid dehydrogenase complexes. Involved in the hyperactivation of spermatazoa during capacitation and in the spermatazoal acrosome reaction.
|
| GO Classification
|
| Biological Process |
| lysine catabolic process |
| tricarboxylic acid cycle |
| proteolysis |
| aging |
| cell redox homeostasis |
| acetyl-CoA biosynthetic process from pyruvate |
| dihydrolipoamide metabolic process |
| gastrulation |
| lipoate metabolic process |
| mitochondrial electron transport, NADH to ubiquinone |
| regulation of acetyl-CoA biosynthetic process from pyruvate |
| regulation of membrane potential |
| sperm capacitation |
| branched-chain amino acid catabolic process |
| cellular nitrogen compound metabolic process |
| pyruvate metabolic process |
| 2-oxoglutarate metabolic process |
| Cellular Component |
| mitochondrial matrix |
| nucleus |
| flagellum |
| acrosomal matrix |
| oxoglutarate dehydrogenase complex |
| pyruvate dehydrogenase complex |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| oxidoreductase activity |
| oxidoreductase activity, acting on nadh or nadph |
| oxidoreductase activity, acting on a sulfur group of donors, nad or nadp as acceptor |
| dihydrolipoyl dehydrogenase activity |
| fad or fadh2 binding |
| binding |
| catalytic activity |
| Molecular Function |
| NAD binding |
| flavin adenine dinucleotide binding |
| dihydrolipoyl dehydrogenase activity |
| lipoamide binding |
| Process |
| cell redox homeostasis |
| metabolic process |
| cellular process |
| oxidation reduction |
| cellular homeostasis |
|
| Cellular Location
|
- Mitochondrion matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Glycolysis / Gluconeogenesis | Not Available |  | | Citrate cycle (TCA cycle) | Not Available |  | | Glycine, serine and threonine metabolism | Not Available |  | | Citric Acid Cycle | 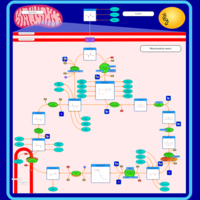 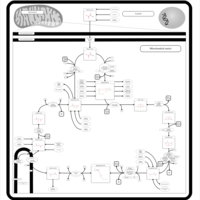 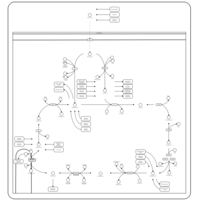 |  | | Congenital lactic acidosis | 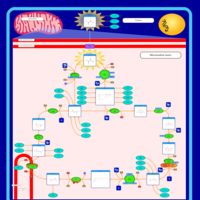 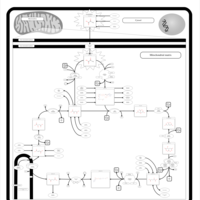 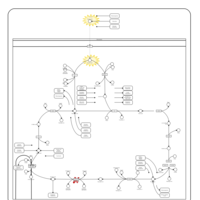 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 7 |
| Locus
| 7q31-q32 |
| SNPs
| DLD |
| Gene Sequence
|
>1530 bp
ATGCAGAGCTGGAGTCGTGTGTACTGCTCCTTGGCCAAGAGAGGCCATTTCAATCGAATA
TCTCATGGCCTACAGGGACTTTCTGCAGTGCCTCTGAGAACTTACGCAGATCAGCCGATT
GATGCTGATGTAACAGTTATAGGTTCTGGTCCTGGAGGATATGTTGCTGCTATTAAAGCT
GCCCAGTTAGGCTTCAAGACAGTCTGCATTGAGAAAAATGAAACACTTGGTGGAACATGC
TTGAATGTTGGTTGTATTCCTTCTAAGGCTTTATTGAACAACTCTCATTATTACCATATG
GCCCATGGAAAAGATTTTGCATCTAGAGGAATTGAAATGTCCGAAGTTCGCTTGAATTTA
GACAAGATGATGGAGCAGAAGAGTACTGCAGTAAAAGCTTTAACAGGTGGAATTGCCCAC
TTATTCAAACAGAATAAGGTTGTTCATGTCAATGGATATGGAAAGATAACTGGCAAAAAT
CAAGTCACTGCTACGAAAGCTGATGGCGGCACTCAGGTTATTGATACAAAGAACATTCTT
ATAGCCACGGGTTCAGAAGTTACTCCTTTTCCTGGAATCACGATAGATGAAGATACAATA
GTGTCATCTACAGGTGCTTTATCTTTAAAAAAAGTTCCAGAAAAGATGGTTGTTATTGGT
GCAGGAGTAATAGGTGTAGAATTGGGTTCAGTTTGGCAAAGACTTGGTGCAGATGTGACA
GCAGTTGAATTTTTAGGTCATGTAGGTGGAGTTGGAATTGATATGGAGATATCTAAAAAC
TTTCAACGCATCCTTCAAAAACAGGGGTTTAAATTTAAATTGAATACAAAGGTTACTGGT
GCTACCAAGAAGTCAGATGGAAAAATTGATGTTTCTATTGAAGCTGCTTCTGGTGGTAAA
GCTGAAGTTATCACTTGTGATGTACTCTTGGTTTGCATTGGCCGACGACCCTTTACTAAG
AATTTGGGACTAGAAGAGCTGGGAATTGAACTAGATCCCAGAGGTAGAATTCCAGTCAAT
ACCAGATTTCAAACTAAAATTCCAAATATCTATGCCATTGGTGATGTAGTTGCTGGTCCA
ATGCTGGCTCACAAAGCAGAGGATGAAGGCATTATCTGTGTTGAAGGAATGGCTGGTGGT
GCTGTGCACATTGACTACAATTGTGTGCCATCAGTGATTTACACACACCCTGAAGTTGCT
TGGGTTGGCAAATCAGAAGAGCAGTTGAAAGAAGAGGGTATTGAGTACAAAGTTGGGAAA
TTCCCATTTGCTGCTAACAGCAGAGCTAAGACAAATGCTGACACAGATGGCATGGTGAAG
ATCCTTGGGCAGAAATCGACAGACAGAGTACTGGGAGCACATATTCTTGGACCAGGTGCT
GGAGAAATGGTAAATGAAGCTGCTCTTGCTTTGGAATATGGAGCATCCTGTGAAGATATA
GCTAGAGTCTGTCATGCACATCCGACCTTATCAGAAGCTTTTAGAGAAGCAAATCTTGCT
GCGTCATTTGGCAAATCAATCAACTTTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 509 |
| Molecular Weight
| 54176.91 |
| Theoretical pI
| 7.855 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Dihydrolipoyl dehydrogenase, mitochondrial
MQSWSRVYCSLAKRGHFNRISHGLQGLSAVPLRTYADQPIDADVTVIGSGPGGYVAAIKA
AQLGFKTVCIEKNETLGGTCLNVGCIPSKALLNNSHYYHMAHGKDFASRGIEMSEVRLNL
DKMMEQKSTAVKALTGGIAHLFKQNKVVHVNGYGKITGKNQVTATKADGGTQVIDTKNIL
IATGSEVTPFPGITIDEDTIVSSTGALSLKKVPEKMVVIGAGVIGVELGSVWQRLGADVT
AVEFLGHVGGVGIDMEISKNFQRILQKQGFKFKLNTKVTGATKKSDGKIDVSIEAASGGK
AEVITCDVLLVCIGRRPFTKNLGLEELGIELDPRGRIPVNTRFQTKIPNIYAIGDVVAGP
MLAHKAEDEGIICVEGMAGGAVHIDYNCVPSVIYTHPEVAWVGKSEEQLKEEGIEYKVGK
FPFAANSRAKTNADTDGMVKILGQKSTDRVLGAHILGPGAGEMVNEAALALEYGASCEDI
ARVCHAHPTLSEAFREANLAASFGKSINF
|
| External Links |
|---|
| GenBank ID Protein
| 91199540 |
| UniProtKB/Swiss-Prot ID
| P09622 |
| UniProtKB/Swiss-Prot Entry Name
| DLDH_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| NM_000108.3 |
| GeneCard ID
| DLD |
| GenAtlas ID
| DLD |
| HGNC ID
| HGNC:2898 |
| References |
|---|
| General References
| Not Available |


