| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:47:06 UTC |
|---|
| Updated at | 2020-11-18 16:38:56 UTC |
|---|
| CannabisDB ID | CDB004887 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | 5'-Methylthioadenosine |
|---|
| Description | 5'-Methylthioadenosine, also known as MTA or thiomethyladenosine, belongs to the class of organic compounds known as 5'-deoxy-5'-thionucleosides. These are 5'-deoxyribonucleosides in which the ribose is thio-substituted at the 5'position by a S-alkyl group. 5'-Methylthioadenosine is a strong basic compound (based on its pKa). 5'-Methylthioadenosine exists in all living species, ranging from bacteria to humans. Within humans, 5'-methylthioadenosine participates in a number of enzymatic reactions. In particular, 5'-methylthioadenosine and spermidine can be biosynthesized from S-adenosylmethioninamine and putrescine through its interaction with the enzyme spermidine synthase. In addition, 5'-methylthioadenosine can be converted into 5-methylthioribose 1-phosphate and L-methionine; which is catalyzed by the enzyme S-methyl-5'-thioadenosine phosphorylase. In humans, 5'-methylthioadenosine is involved in the metabolic disorder called hypermethioninemia. Outside of the human body, 5'-Methylthioadenosine has been detected, but not quantified in, several different foods, such as chia, black elderberries, kumquats, jew's ears, and pine nuts. This could make 5'-methylthioadenosine a potential biomarker for the consumption of these foods. Adenosine with the hydroxy group at C-5' substituted with a methylthio (methylsulfanyl) group. 5'-Methylthioadenosine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 5'-Deoxy-5'-(methylthio)adenosine | ChEBI | | 5-Methylthioadenosine | ChEBI | | 9-(5-S-Methyl-5-thio-beta-D-ribofuranosyl)-9H-purin-6-amine | ChEBI | | Methylthioadenosine | ChEBI | | MTA | ChEBI | | S-Methyl-5'-thioadenosine | ChEBI | | Thiomethyladenosine | ChEBI | | 9-(5-S-Methyl-5-thio-b-D-ribofuranosyl)-9H-purin-6-amine | Generator | | 9-(5-S-Methyl-5-thio-β-D-ribofuranosyl)-9H-purin-6-amine | Generator | | 1-(6-Amino-9H-purin-9-yl)-1-deoxy-5-S-methyl-5-thio-beta-D-ribofuranose | HMDB | | 1-(6-Amino-9H-purin-9-yl)-1-deoxy-5-S-methyl-5-thio-beta-delta-ribofuranose | HMDB | | 5'-(Methylthio)-5'-deoxyadenosine | HMDB | | 5'-(Methylthio)adenosine | HMDB | | 5'-S-Methyl-5'-thio-adenosine | HMDB | | 5'-S-Methyl-5'-thioadenosine | HMDB | | S-Methyl-5-thioadenosine | HMDB | | Adenine(5'-deoxy-5'-methylthio)9-beta-D-furanoriboside | HMDB | | 5-MTDA | HMDB | | 5'-Methylthio-5'-deoxyadenosine | HMDB | | 5'-Deoxy-5'-methylthioadenosine | HMDB | | 5'-Methylthioadenosine, methyl-(14)C-labeled | HMDB | | 5'-Methylthioadenosine | ChEBI |
|
|---|
| Chemical Formula | C11H15N5O3S |
|---|
| Average Molecular Weight | 297.33 |
|---|
| Monoisotopic Molecular Weight | 297.0896 |
|---|
| IUPAC Name | (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-[(methylsulfanyl)methyl]oxolane-3,4-diol |
|---|
| Traditional Name | methylthioadenosine |
|---|
| CAS Registry Number | 2457-80-9 |
|---|
| SMILES | CSC[C@H]1O[C@H]([C@H](O)[C@@H]1O)N1C=NC2=C1N=CN=C2N |
|---|
| InChI Identifier | InChI=1S/C11H15N5O3S/c1-20-2-5-7(17)8(18)11(19-5)16-4-15-6-9(12)13-3-14-10(6)16/h3-5,7-8,11,17-18H,2H2,1H3,(H2,12,13,14)/t5-,7-,8-,11-/m1/s1 |
|---|
| InChI Key | WUUGFSXJNOTRMR-IOSLPCCCSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as 5'-deoxy-5'-thionucleosides. These are 5'-deoxyribonucleosides in which the ribose is thio-substituted at the 5'position by a S-alkyl group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Nucleosides, nucleotides, and analogues |
|---|
| Class | 5'-deoxyribonucleosides |
|---|
| Sub Class | 5'-deoxy-5'-thionucleosides |
|---|
| Direct Parent | 5'-deoxy-5'-thionucleosides |
|---|
| Alternative Parents | |
|---|
| Substituents | - 5'-deoxy-5'-thionucleoside
- Glycosyl compound
- N-glycosyl compound
- 6-aminopurine
- Pentose monosaccharide
- Imidazopyrimidine
- Purine
- Aminopyrimidine
- Monosaccharide
- N-substituted imidazole
- Pyrimidine
- Imidolactam
- Azole
- Tetrahydrofuran
- Imidazole
- Heteroaromatic compound
- Secondary alcohol
- 1,2-diol
- Organoheterocyclic compound
- Dialkylthioether
- Sulfenyl compound
- Thioether
- Azacycle
- Oxacycle
- Organic oxygen compound
- Alcohol
- Hydrocarbon derivative
- Organopnictogen compound
- Amine
- Primary amine
- Organic nitrogen compound
- Organonitrogen compound
- Organooxygen compound
- Organosulfur compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | 5'-Methylthioadenosine, non-derivatized, GC-MS Spectrum | splash10-000i-0940000000-16b1106966ac98c7a107 | Spectrum | | GC-MS | 5'-Methylthioadenosine, non-derivatized, GC-MS Spectrum | splash10-000i-0940000000-16b1106966ac98c7a107 | Spectrum | | GC-MS | 5'-Methylthioadenosine, non-derivatized, GC-MS Spectrum | splash10-000i-0940000000-30fc68e7b5ff7576069b | Spectrum | | GC-MS | 5'-Methylthioadenosine, non-derivatized, GC-MS Spectrum | splash10-01ti-0900000000-312bd8eec3c461e919f9 | Spectrum | | Predicted GC-MS | 5'-Methylthioadenosine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-056r-9530000000-5fbf6b1a856ad3827980 | Spectrum | | Predicted GC-MS | 5'-Methylthioadenosine, 2 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0h70-9636300000-67e425eba97fe420c464 | Spectrum | | Predicted GC-MS | 5'-Methylthioadenosine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | 5'-Methylthioadenosine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000j-0940000000-55655e7a0a6c8acba0f0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-000i-1900000000-f37865ca2029a623ace7 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-000i-2900000000-ae0e3df9bf25c100940d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0002-0090000000-72139812c5b2ede077ed | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-000i-0910000000-0c3d173f36272e3271eb | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-000i-0900000000-f3443ca133811f52c5d7 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-000i-1900000000-49cc299525d7fca27818 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-000i-3900000000-39bebdcaa7ce2695465b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-000i-0900000000-76b935f4f0a78c15ca0e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT (LC/MSD Trap XCT, Agilent Technologies) , Positive | splash10-0002-4900000000-d8c0df43305b934d71dc | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-000i-0900000000-d3bfbeb6fb95e3768ef6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-001i-0910000000-56784be58ef9065f21f6 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-001i-0910000000-56784be58ef9065f21f6 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-0002-0090000000-72139812c5b2ede077ed | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-000i-0910000000-0c3d173f36272e3271eb | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-000i-0900000000-f3443ca133811f52c5d7 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-000i-1900000000-6ec86a11bf8ad7331bd2 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , positive | splash10-000i-3900000000-39bebdcaa7ce2695465b | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-000i-0900000000-b9006ef8eecdaefc89a6 | 2017-09-14 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-000i-0940000000-eb168427f389d7079abf | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-000i-0900000000-714e20a3376683008e65 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-000i-2900000000-cb0adab85341c49ced5b | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9220000000-14da12fd68bc6d33eed9 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-4910000000-324b5daf65bdaa0d2bdb | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-053r-4900000000-14e9c89fcdd488f204aa | 2016-09-12 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Methionine Metabolism |    |  | | Spermidine and Spermine Biosynthesis |  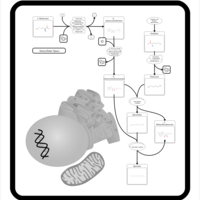 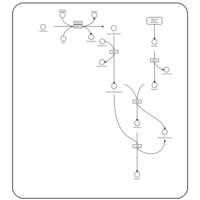 | Not Available | | Cystathionine Beta-Synthase Deficiency | 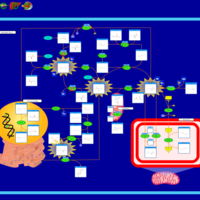 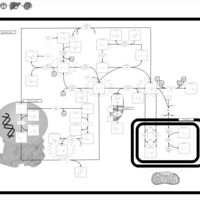 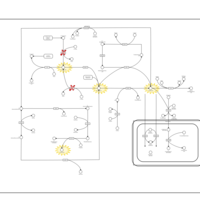 | Not Available | | Hypermethioninemia | 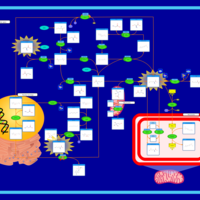 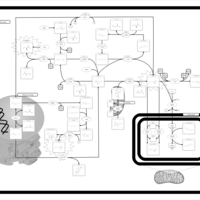 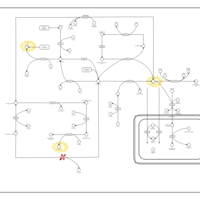 | Not Available | | S-Adenosylhomocysteine (SAH) Hydrolase Deficiency | 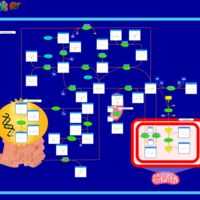 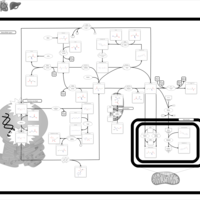 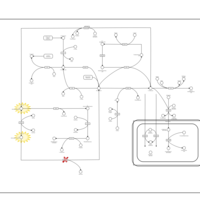 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0001173 |
|---|
| DrugBank ID | DB02282 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB031156 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 388321 |
|---|
| KEGG Compound ID | C00170 |
|---|
| BioCyc ID | 5-METHYLTHIOADENOSINE |
|---|
| BiGG ID | 34127 |
|---|
| Wikipedia Link | Not Available |
|---|
| METLIN ID | 3425 |
|---|
| PubChem Compound | 439176 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17509 |
|---|
| References |
|---|
| General References | Not Available |
|---|
