| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 18:39:43 UTC |
|---|
| Updated at | 2021-01-22 17:44:17 UTC |
|---|
| CannabisDB ID | CDB004816 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | L-Lysine |
|---|
| Description | Lysin and its bioctive form L-lysine is an essential amino acid. Normal human requirements for lysine have been found to be about 8 g per day or 12 mg/kg in adults. Children and infants need more, 44 mg/kg per day for an eleven to-twelve-year old, and 97 mg/kg per day for three-to six-month old. Lysine is highly concentrated in muscle compared to most other amino acids. Lysine is high in foods such as wheat germ, cottage cheese and chicken. Of meat products, wild game and pork have the highest concentration of lysine. Fruits and vegetables contain little lysine, except avocados. Normal lysine metabolism is dependent upon many nutrients including niacin, vitamin B6, riboflavin, vitamin C, glutamic acid and iron. Excess arginine antagonizes lysine. Several inborn errors of lysine metabolism are known, such as cystinuria, hyperdibasic aminoaciduria I, lysinuric protein intolerance, propionic acidemia, and tyrosinemia I. Most are marked by mental retardation with occasional diverse symptoms such as absence of secondary sex characteristics, undescended testes, abnormal facial structure, anemia, obesity, enlarged liver and spleen, and eye muscle imbalance. Lysine also may be a useful adjunct in the treatment of osteoporosis. Although high protein diets result in loss of large amounts of calcium in urine, so does lysine deficiency. Lysine may be an adjunct therapy because it reduces calcium losses in urine. Lysine deficiency also may result in immunodeficiency. Requirements for this amino acid are probably increased by stress. Lysine toxicity has not occurred with oral doses in humans. Lysine dosages are presently too small and may fail to reach the concentrations necessary to prove potential therapeutic applications. Lysine metabolites, amino caproic acid and carnitine have already shown their therapeutic potential. Thirty grams daily of amino caproic acid has been used as an initial daily dose in treating blood clotting disorders, indicating that the proper doses of lysine, its precursor, have yet to be used in medicine. As a widespread aminoacid in nature, L-lysine is also present in the Cannabis sativa plant (PMID: 6991645 ) and it has also been detected in the smoke produced through the combustion of Cannabis (Ref: Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| (S)-2,6-Diaminohexanoic acid | ChEBI | | (S)-alpha,epsilon-Diaminocaproic acid | ChEBI | | (S)-Lysine | ChEBI | | 6-Ammonio-L-norleucine | ChEBI | | K | ChEBI | | L-2,6-Diaminocaproic acid | ChEBI | | L-Lysin | ChEBI | | Lys | ChEBI | | Lysina | ChEBI | | Lysine | ChEBI | | Lysine acid | ChEBI | | Lysinum | ChEBI | | 2,6-Diaminohexanoic acid | Kegg | | (S)-2,6-Diaminohexanoate | Generator | | (S)-a,epsilon-Diaminocaproate | Generator | | (S)-a,epsilon-Diaminocaproic acid | Generator | | (S)-alpha,epsilon-Diaminocaproate | Generator | | (S)-Α,epsilon-diaminocaproate | Generator | | (S)-Α,epsilon-diaminocaproic acid | Generator | | L-2,6-Diaminocaproate | Generator | | 2,6-Diaminohexanoate | Generator | | (+)-S-Lysine | HMDB | | (S)-2,6-Diamino-hexanoate | HMDB | | (S)-2,6-Diamino-hexanoic acid | HMDB | | (S)-a,e-Diaminocaproate | HMDB | | (S)-a,e-Diaminocaproic acid | HMDB | | 6-Amino-aminutrin | HMDB | | 6-Amino-L-norleucine | HMDB | | a-Lysine | HMDB | | alpha-Lysine | HMDB | | Aminutrin | HMDB | | H-Lys-OH | HMDB | | L-(+)-Lysine | HMDB | | L-2,6-Diainohexanoate | HMDB | | L-2,6-Diainohexanoic acid | HMDB | | L-Lys | HMDB | | Acetate, lysine | HMDB | | Enisyl | HMDB | | Lysine hydrochloride | HMDB | | L Lysine | HMDB | | Lysine acetate | HMDB |
|
|---|
| Chemical Formula | C6H14N2O2 |
|---|
| Average Molecular Weight | 146.19 |
|---|
| Monoisotopic Molecular Weight | 146.1055 |
|---|
| IUPAC Name | (2S)-2,6-diaminohexanoic acid |
|---|
| Traditional Name | L-lysine |
|---|
| CAS Registry Number | 56-87-1 |
|---|
| SMILES | NCCCC[C@H](N)C(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h5H,1-4,7-8H2,(H,9,10)/t5-/m0/s1 |
|---|
| InChI Key | KDXKERNSBIXSRK-YFKPBYRVSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
| Class | Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
| Direct Parent | L-alpha-amino acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - L-alpha-amino acid
- Medium-chain fatty acid
- Amino fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Organic nitrogen compound
- Amine
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 224.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000 mg/mL | Not Available | | logP | -3.05 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| GC-MS | L-Lysine, 3 TMS, GC-MS Spectrum | splash10-00di-3910000000-98c565675de67aa87900 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0ab9-1910000000-87ef8534f592041f50f2 | Spectrum | | GC-MS | L-Lysine, 4 TMS, GC-MS Spectrum | splash10-0a4i-1921000000-84f7815b0f650fa17444 | Spectrum | | GC-MS | L-Lysine, 3 TMS, GC-MS Spectrum | splash10-001i-9600000000-823408dba509cb204acf | Spectrum | | GC-MS | L-Lysine, 3 TMS, GC-MS Spectrum | splash10-00di-3910000000-4f5578af5e7d8b6c49f7 | Spectrum | | GC-MS | L-Lysine, 4 TMS, GC-MS Spectrum | splash10-0adi-1921000000-4e56d95e623e792f9e6b | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-00di-0921000000-eeb49e57bc1a75193058 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0ab9-0921000000-ebb902be0f3754225b2f | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-00di-3910000000-98c565675de67aa87900 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0ab9-1910000000-87ef8534f592041f50f2 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0fdk-3923000000-15b84c2649c1b0455de1 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0a4i-1921000000-84f7815b0f650fa17444 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-001i-9600000000-823408dba509cb204acf | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-00di-3910000000-4f5578af5e7d8b6c49f7 | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0adi-1921000000-4e56d95e623e792f9e6b | Spectrum | | GC-MS | L-Lysine, non-derivatized, GC-MS Spectrum | splash10-0abi-1900000000-9ad174122e4d6e003eb8 | Spectrum | | Predicted GC-MS | L-Lysine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0089-9100000000-974cc55c9130ed5213eb | Spectrum | | Predicted GC-MS | L-Lysine, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0ue9-9700000000-57b24ae819b6ec26bfd2 | Spectrum | | Predicted GC-MS | L-Lysine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Lysine, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Lysine, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Lysine, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Lysine, TBDMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | L-Lysine, TBDMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003r-8900000000-470a0beb4f338ed89bca | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-74e9193d9d33c2509bfa | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-822c4e78250fffa56e39 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-04f9a62a77fb5a37ca22 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-035035ecfa084671479b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-5bb15839f86f4fca0d0b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6c8ef03aa83eb1cab35b | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-01ot-0910000000-c182a7dcdbc260666978 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4j-0900000000-6c5f378cef2f14204e15 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-41e1a6499097748934b0 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0390000000-7270a0b85b9e3f9f6373 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-a997e809874357908880 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-2900000000-f64110414f82c93e1fe8 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-014m-9200000000-33a38e6370811c5ddc82 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-3a8b0b6e62f5c66d3720 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-d5167570d11d77fd541e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0002-0900000000-a46231bd529176101129 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-001i-9200000000-f8b9f01b2a9886c51df9 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-001i-9000000000-ace0361476d939043e98 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-001i-9000000000-5b06b6e9dcf9faf8e89c | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-053r-9000000000-2894ef6d7f72ea71688e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-0002-0900000000-290902f43cf851e8ef5e | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-f81193f6b50235ec8147 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-8931d2193adab166d5e4 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0002-0900000000-b4825cc64fcb830c6967 | 2012-08-31 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Transcription/Translation | Not Available | Not Available | | Lysine Degradation | 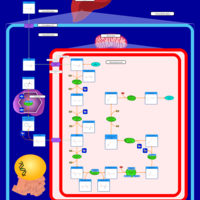 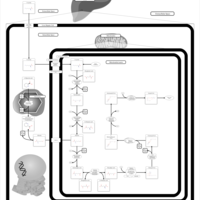 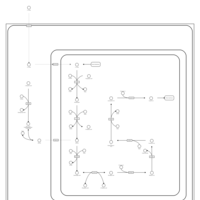 |  | | Biotin Metabolism | 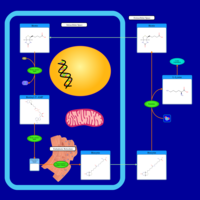   | 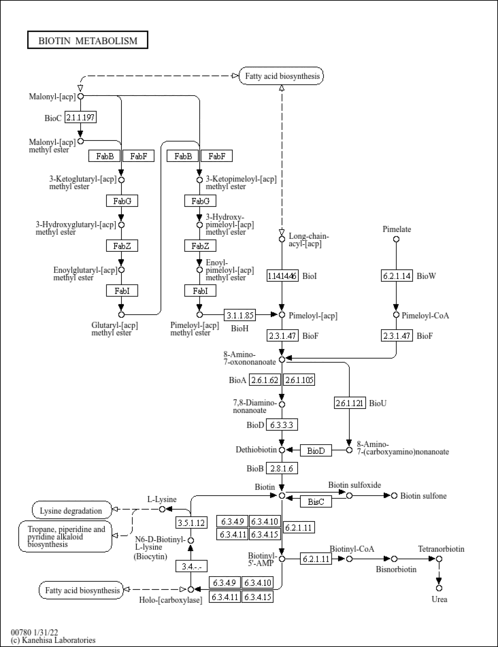 | | Carnitine Synthesis |  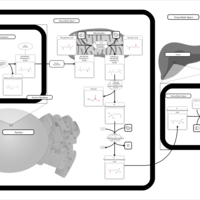 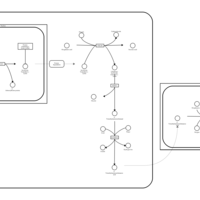 | Not Available | | Biotinidase Deficiency | 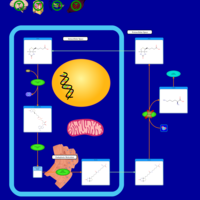 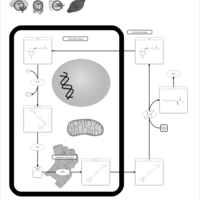  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-lysine N-methyltransferase EZH2 | EZH2 | 7q35-q36 | Q15910 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details |
|
|---|
| Transcriptional Factors | |
| Histone-lysine N-methyltransferase, H3 lysine-79 specific | DOT1L | 19p13.3 | Q8TEK3 | details | | Histone-lysine N-methyltransferase SETD7 | SETD7 | 4q28 | Q8WTS6 | details | | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific | NSD1 | 5q35 | Q96L73 | details | | Histone-lysine N-methyltransferase SETDB1 | SETDB1 | 1q21 | Q15047 | details | | Histone-lysine N-methyltransferase EHMT2 | EHMT2 | 6p21.31 | Q96KQ7 | details | | Histone-lysine N-methyltransferase EHMT1 | EHMT1 | 9q34.3 | Q9H9B1 | details | | PR domain zinc finger protein 2 | PRDM2 | 1p36.21 | Q13029 | details | | Histone-lysine N-methyltransferase MLL | MLL | 11q23 | Q03164 | details | | Histone-lysine N-methyltransferase MLL3 | MLL3 | 7q36.1 | Q8NEZ4 | details | | Histone-lysine N-methyltransferase EZH2 | EZH2 | 7q35-q36 | Q15910 | details | | Histone-lysine N-methyltransferase MLL2 | MLL2 | 12q13.12 | O14686 | details | | Histone-lysine N-methyltransferase MLL4 | WBP7 | 19q13.1 | Q9UMN6 | details | | Histone-lysine N-methyltransferase NSD2 | WHSC1 | 4p16.3 | O96028 | details | | Histone-lysine N-methyltransferase NSD3 | WHSC1L1 | 8p11.2 | Q9BZ95 | details | | Histone-lysine N-methyltransferase SETD2 | SETD2 | 3p21.31 | Q9BYW2 | details | | Lysine-specific demethylase 2A | KDM2A | 11q13.2 | Q9Y2K7 | details | | Lysine-specific demethylase 2B | KDM2B | 12q24.31 | Q8NHM5 | details |
|
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000182 |
|---|
| DrugBank ID | DB00123 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB000474 |
|---|
| KNApSAcK ID | C00001378 |
|---|
| Chemspider ID | 5747 |
|---|
| KEGG Compound ID | C00047 |
|---|
| BioCyc ID | LYS |
|---|
| BiGG ID | 33655 |
|---|
| Wikipedia Link | Lysine |
|---|
| METLIN ID | 5200 |
|---|
| PubChem Compound | 5962 |
|---|
| PDB ID | LYS |
|---|
| ChEBI ID | 18019 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|

