| Identification |
|---|
| HMDB Protein ID
| CDBP00422 |
| Secondary Accession Numbers
| Not Available |
| Name
| Inosine triphosphate pyrophosphatase |
| Description
| Not Available |
| Synonyms
|
- ITPase
- Inosine triphosphatase
- Putative oncogene protein hlc14-06-p
- Non-canonical purine NTP pyrophosphatase
- Non-standard purine NTP pyrophosphatase
- Nucleoside-triphosphate diphosphatase
- Nucleoside-triphosphate pyrophosphatase
- NTPase
|
| Gene Name
| ITPA |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity |
| Specific Function
| Pyrophosphatase that hydrolyzes the non-canonical purine nucleotides inosine triphosphate (ITP), deoxyinosine triphosphate (dITP) as well as 2'-deoxy-N-6-hydroxylaminopurine triposphate (dHAPTP) and xanthosine 5'-triphosphate (XTP) to their respective monophosphate derivatives. The enzyme does not distinguish between the deoxy- and ribose forms. Probably excludes non-canonical purines from RNA and DNA precursor pools, thus preventing their incorporation into RNA and DNA and avoiding chromosomal lesions.
|
| GO Classification
|
| Biological Process |
| chromosome organization |
| ITP catabolic process |
| Cellular Component |
| cytoplasm |
| Function |
| catalytic activity |
| hydrolase activity |
| Molecular Function |
| metal ion binding |
| dITP diphosphatase activity |
| nucleotide binding |
|
| Cellular Location
|
- Cytoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Drug metabolism - other enzymes | Not Available | 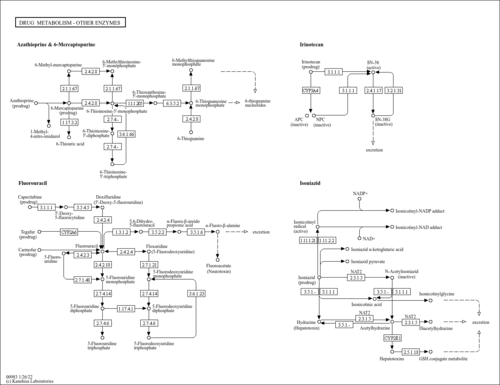 | | Purine Metabolism |    |  | | Adenosine Deaminase Deficiency |   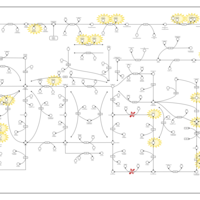 | Not Available | | Adenylosuccinate Lyase Deficiency | 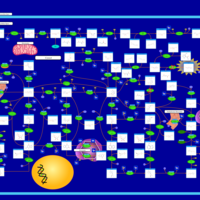 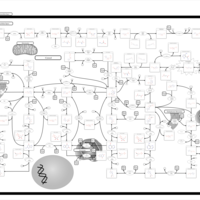 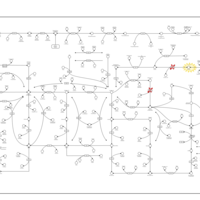 | Not Available | | Gout or Kelley-Seegmiller Syndrome | 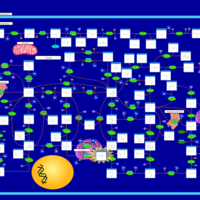 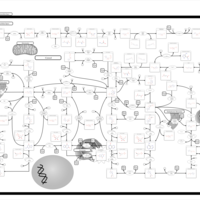 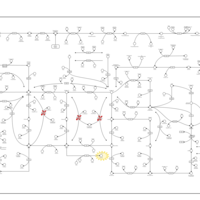 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 20 |
| Locus
| 20p |
| SNPs
| ITPA |
| Gene Sequence
|
>585 bp
ATGGCGGCCTCATTGGTGGGGAAGAAGATCGTGTTTGTAACGGGGAACGCCAAGAAGCTG
GAGGAGGTCGTTCAGATTCTAGGAGATAAGTTTCCATGCACTTTGGTGGCACAGAAAATT
GACCTGCCGGAGTACCAAGGGGAGCCGGATGAGATTTCCATACAGAAATGTCAGGAGGCA
GTTCGCCAGGTACAGGGGCCCGTGCTGGTTGAGGACACTTGTCTGTGCTTCAATGCCCTT
GGAGGGCTCCCCGGCCCCTACATAAAGTGGTTTCTGGAGAAGTTAAAGCCTGAAGGTCTC
CACCAGCTCCTGGCCGGGTTCGAGGACAAGTCAGCCTATGCGCTCTGCACGTTTGCACTC
AGCACCGGGGACCCAAGCCAGCCCGTGCGCCTGTTCAGGGGCCGGACCTCGGGCCGGATC
GTGGCACCCAGAGGCTGCCAGGACTTTGGCTGGGACCCCTGCTTTCAGCCTGATGGATAT
GAGCAGACGTACGCAGAGATGCCTAAGGCGGAGAAGAACGCTGTCTCCCATCGCTTCCGG
GCCCTGCTGGAGCTGCAGGAATACTTTGGCAGTTTGGCAGCTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 194 |
| Molecular Weight
| 16833.23 |
| Theoretical pI
| 7.174 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Inosine triphosphate pyrophosphatase
MAASLVGKKIVFVTGNAKKLEEVVQILGDKFPCTLVAQKIDLPEYQGEPDEISIQKCQEA
VRQVQGPVLVEDTCLCFNALGGLPGPYIKWFLEKLKPEGLHQLLAGFEDKSAYALCTFAL
STGDPSQPVRLFRGRTSGRIVAPRGCQDFGWDPCFQPDGYEQTYAEMPKAEKNAVSHRFR
ALLELQEYFGSLAA
|
| External Links |
|---|
| GenBank ID Protein
| 21104378 |
| UniProtKB/Swiss-Prot ID
| Q9BY32 |
| UniProtKB/Swiss-Prot Entry Name
| ITPA_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AB062127 |
| GeneCard ID
| ITPA |
| GenAtlas ID
| ITPA |
| HGNC ID
| HGNC:6176 |
| References |
|---|
| General References
| Not Available |

