| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:22:15 UTC |
|---|
| Updated at | 2020-12-07 19:06:57 UTC |
|---|
| CannabisDB ID | CDB006165 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Maltose |
|---|
| Description | D-Maltose, also known as malt sugar, maltobiose or alpha-malt sugar, is a disaccharide consisting of two glucose subunits joined via an alpha-1-4 bond. Carbohydrates are generally divided into different categories based on the number of sugar subunits; monosaccharides (1 sugar subunit), disaccharides (2 sugar subunits), oligosaccharides (3 or more sugar subunits) and polysaccharides (hundreds to thousands of subunits, including starch). Maltose is a common byproduct of starch. When beta-amylase breaks down starch, it removes two glucose units at a time, thereby producing maltose. Maltose can be broken down to glucose by the maltase enzyme, which catalyses the hydrolysis of the glycosidic bond. D-Maltose exists in all living species, ranging from bacteria to humans. Maltose was 'discovered' by Augustin-Pierre Dubrunfaut, although this discovery was not widely accepted until it was confirmed in 1872 by Irish chemist and brewer Cornelius O'Sullivan. Its name comes from malt, combined with the suffix '-ose' which is used in naming all sugars. Like glucose, maltose is a reducing sugar, because the ring of one of the two glucose units can open to present a free aldehyde group. Maltose, when placed in aqueous solution, exhibits mutarotation. This is because the α and β isomers that are formed by the different conformations of the anomeric carbon have different specific rotations. In aqueous solutions, these two forms are in equilibrium. Maltose has a sweet taste, but is only about 30-60% as sweet as sucrose. Within humans, D-maltose participates in a number of enzymatic reactions. In particular, D-maltose can be converted into Alpha-D-glucose; which is mediated by intestinal maltase-glucoamylase. In addition, D-maltose can be converted into Alpha-D-glucose through its interaction with the enzyme glycogen debranching enzyme. In humans, D-maltose is involved in the metabolic disorder called the sucrase-isomaltase deficiency. Outside of the human body, D-maltose is found in high concentrations a variety of foods such as fruit gums, marshmallows, and oriental wheats. It is found in lower concentrations in bread, bagels, breakfast cereals, crackers, cornmeal, wheat, barley, candies, honey, molasses, beer and wines. Maltose is found mainly in grains and cereals. Wheat, corn, barley and rye all contain varying amounts of maltose. For some foods, cooking can increase the maltose content. For example, raw sweet potatoes don't have any maltose, but cooked sweet potatoes contain maltose at varying amounts depending on the type of sweet potato. Maltose is a component of malt, a substance which is obtained in the process of allowing grain to soften in water and germinate. Maltose is also found in partially hydrolysed starch products like maltodextrin, corn syrup and acid-thinned starch. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 4-O-alpha-D-Glucopyranosyl-alpha-D-mannopyranose | ChEBI | | 4-O-a-D-Glucopyranosyl-a-D-mannopyranose | Generator | | 4-O-Α-D-glucopyranosyl-α-D-mannopyranose | Generator | | 1-alpha-D-Glucopyranosyl-4-alpha-D-glucopyranose | HMDB | | 1-alpha-delta-Glucopyranosyl-4-alpha-delta-glucopyranose | HMDB | | 4-(alpha-D-Glucopyranosido)-alpha-glucopyranose | HMDB | | 4-(alpha-D-Glucosido)-D-glucose | HMDB | | 4-(alpha-delta-Glucopyranosido)-alpha-glucopyranose | HMDB | | 4-(alpha-delta-Glucosido)-delta-glucose | HMDB | | 4-O-a-D-Glucopyranosyl-D-glucose | HMDB | | 4-O-alpha-D-Glucopyranosyl-D-glucopyranose | HMDB | | 4-O-alpha-D-Glucopyranosyl-D-glucose | HMDB | | 4-O-alpha-delta-Glucopyranosyl-delta-glucopyranose | HMDB | | 4-O-alpha-delta-Glucopyranosyl-delta-glucose | HMDB | | Advantose 100 | HMDB | | alpha-D-GLCP-(1->4)-D-GLCP | HMDB | | alpha-D-Glucopyranosyl-(1->4)-D-glucopyranose | HMDB | | alpha-D-Glucopyranosyl-(1->4)-D-glucose | HMDB | | alpha-delta-GLCP-(1->4)-delta-GLCP | HMDB | | alpha-delta-Glucopyranosyl-(1->4)-delta-glucopyranose | HMDB | | alpha-delta-Glucopyranosyl-(1->4)-delta-glucose | HMDB | | alpha-Malt sugar | HMDB | | Cextromaltose | HMDB | | D-(+)-Maltose | HMDB | | delta-(+)-Maltose | HMDB | | delta-Maltose | HMDB | | Finetose | HMDB | | Finetose F | HMDB | | Madoros | HMDB | | Malt sugar | HMDB | | Maltobiose | HMDB | | Maltodiose | HMDB | | Maltos | HMDB | | Maltose | HMDB | | Maltose HH | HMDB | | Maltose HHH | HMDB | | Maltose solution | HMDB | | Malzzucker | HMDB | | Martos-10 | HMDB | | Sunmalt | HMDB | | Sunmalt S | HMDB |
|
|---|
| Chemical Formula | C12H22O11 |
|---|
| Average Molecular Weight | 342.3 |
|---|
| Monoisotopic Molecular Weight | 342.1162 |
|---|
| IUPAC Name | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-{[(2R,3S,4R,5S,6S)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}oxane-3,4,5-triol |
|---|
| Traditional Name | (2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-{[(2R,3S,4R,5S,6S)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxy}oxane-3,4,5-triol |
|---|
| CAS Registry Number | 69-79-4 |
|---|
| SMILES | OC[C@H]1O[C@H](O[C@H]2[C@H](O)[C@H](O)[C@@H](O)O[C@@H]2CO)[C@H](O)[C@@H](O)[C@@H]1O |
|---|
| InChI Identifier | InChI=1S/C12H22O11/c13-1-3-5(15)6(16)9(19)12(22-3)23-10-4(2-14)21-11(20)8(18)7(10)17/h3-20H,1-2H2/t3-,4-,5-,6+,7-,8+,9-,10-,11+,12-/m1/s1 |
|---|
| InChI Key | GUBGYTABKSRVRQ-DKBJLJRDSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as o-glycosyl compounds. These are glycoside in which a sugar group is bonded through one carbon to another group via a O-glycosidic bond. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbohydrates and carbohydrate conjugates |
|---|
| Direct Parent | O-glycosyl compounds |
|---|
| Alternative Parents | |
|---|
| Substituents | - O-glycosyl compound
- Disaccharide
- Oxane
- Secondary alcohol
- Hemiacetal
- Oxacycle
- Organoheterocyclic compound
- Polyol
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 102 - 103 °C | Not Available | | Boiling Point | 667.9 °C | Wikipedia | | Water Solubility | 780 mg/mL | Not Available | | logP | Not Available | Not Available |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| Predicted GC-MS | Maltose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-03e9-4897000000-332d470623cd7f1e1ced | Spectrum | | Predicted GC-MS | Maltose, 4 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-014i-3421129000-03a98898fd3278f95502 | Spectrum | | Predicted GC-MS | Maltose, TBDMS_3_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_3_16, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_10, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_11, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_12, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_20, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_32, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TBDMS_4_33, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, "D-Maltose,3TBDMS,#3" TMS, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_5, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_6, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_7, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_1_8, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_2_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_2_2, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_2_3, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Maltose, TMS_2_4, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-01ox-0149000000-74748828832d87932342 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-008c-2950000000-4ebee22b86c2cc9ee3ab | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-00kg-9710000000-041dbc580f99b8881518 | 2012-07-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-03gl-0907000000-a1de7813fbe8b5258c10 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-03e9-0900000000-79cc5059a3f338e8c1dd | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-3900000000-851d0bc84d91b6e09579 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-002f-0749000000-af1d18f2bc3ab8e02883 | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0200-2902000000-67218697344afb9e70fa | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-01td-4900000000-40aa2d8125769803cfbb | 2017-09-01 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0006-0109000000-9d0f9a68a67bafd734ab | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-01rw-3898000000-7f495a1bcfdbed6466c4 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-01ti-9210000000-b8a83657f0266d372bee | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000x-1069000000-9cd867711bca0eecf935 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9365000000-65403961bfb5e35eaa0d | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-000l-9200000000-0de9785c1c5df1430dd2 | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, experimental) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Starch and Sucrose Metabolism | 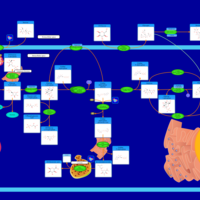 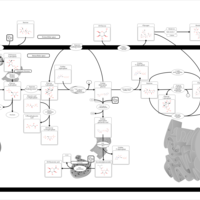 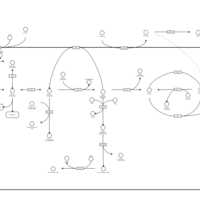 |  | | Glycogen synthetase deficiency | 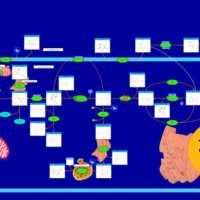 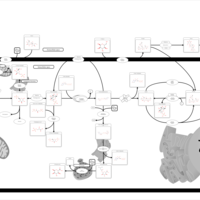 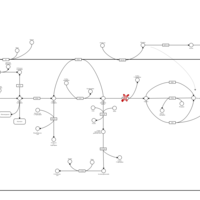 | Not Available | | Glycogenosis, Type III. Cori disease, Debrancher glycogenosis | 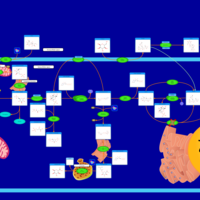 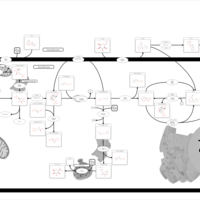 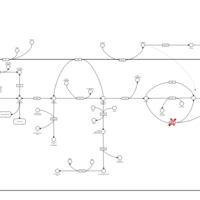 | Not Available | | Glycogenosis, Type IV. Amylopectinosis, Anderson disease | 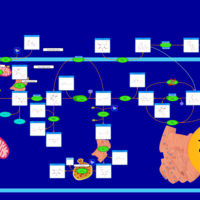 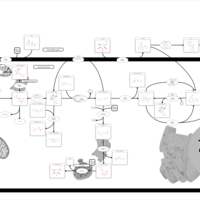 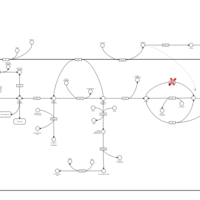 | Not Available | | Glycogenosis, Type VI. Hers disease | 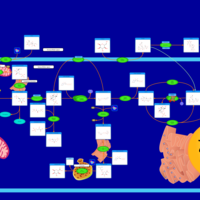 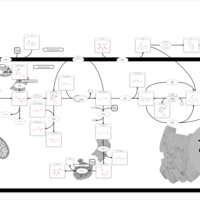  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000163 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001193 |
|---|
| KNApSAcK ID | C00001140 |
|---|
| Chemspider ID | 9166684 |
|---|
| KEGG Compound ID | C00208 |
|---|
| BioCyc ID | Not Available |
|---|
| BiGG ID | 34261 |
|---|
| Wikipedia Link | Maltose |
|---|
| METLIN ID | 413 |
|---|
| PubChem Compound | 10991489 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 47937 |
|---|
| References |
|---|
| General References | Not Available |
|---|
