| Identification |
|---|
| HMDB Protein ID
| CDBP01626 |
| Secondary Accession Numbers
| Not Available |
| Name
| Lysosomal alpha-glucosidase |
| Description
| Not Available |
| Synonyms
|
- 70 kDa lysosomal alpha-glucosidase
- 76 kDa lysosomal alpha-glucosidase
- Acid maltase
- Aglucosidase alfa
|
| Gene Name
| GAA |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Essential for the degradation of glygogen to glucose in lysosomes.
|
| GO Classification
|
| Biological Process |
| ventricular cardiac muscle tissue morphogenesis |
| glycogen catabolic process |
| lysosome organization |
| locomotory behavior |
| neuromuscular process controlling balance |
| muscle cell homeostasis |
| regulation of the force of heart contraction |
| glucose metabolic process |
| cardiac muscle contraction |
| diaphragm contraction |
| neuromuscular process controlling posture |
| maltose metabolic process |
| sucrose metabolic process |
| tongue morphogenesis |
| vacuolar sequestering |
| Cellular Component |
| lysosome |
| lysosomal membrane |
| Function |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on glycosyl bonds |
| hydrolase activity, hydrolyzing o-glycosyl compounds |
| Molecular Function |
| carbohydrate binding |
| alpha-glucosidase activity |
| maltose alpha-glucosidase activity |
| Process |
| metabolic process |
| primary metabolic process |
| carbohydrate metabolic process |
|
| Cellular Location
|
- Lysosome
- Lysosome membrane
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Lysosome | Not Available | 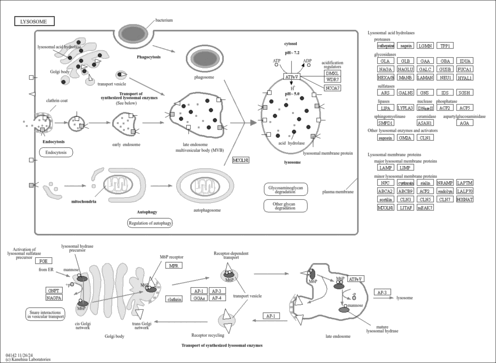 | | Galactose Metabolism |    |  | | Galactosemia | 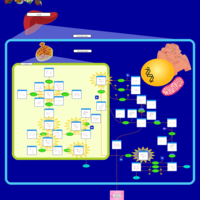 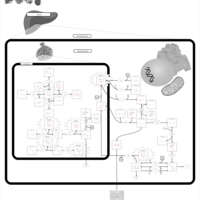 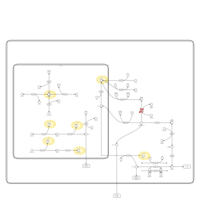 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| Not Available |
| Locus
| Not Available |
| SNPs
| GAA |
| Gene Sequence
|
>2859 bp
ATGGGAGTGAGGCACCCGCCCTGCTCCCACCGGCTCCTGGCCGTCTGCGCCCTCGTGTCC
TTGGCAACCGCTGCACTCCTGGGGCACATCCTACTCCATGATTTCCTGCTGGTTCCCCGA
GAGCTGAGTGGCTCCTCCCCAGTCCTGGAGGAGACTCACCCAGCTCACCAGCAGGGAGCC
AGCAGACCAGGGCCCCGGGATGCCCAGGCACACCCCGGCCGTCCCAGAGCAGTGCCCACA
CAGTGCGACGTCCCCCCCAACAGCCGCTTCGATTGCGCCCCTGACAAGGCCATCACCCAG
GAACAGTGCGAGGCCCGCGGCTGCTGCTACATCCCTGCAAAGCAGGGGCTGCAGGGAGCC
CAGATGGGGCAGCCCTGGTGCTTCTTCCCACCCAGCTACCCCAGCTACAAGCTGGAGAAC
CTGAGCTCCTCTGAAATGGGCTACACGGCCACCCTGACCCGTACCACCCCCACCTTCTTC
CCCAAGGACATCCTGACCCTGCGGCTGGACGTGATGATGGAGACTGAGAACCGCCTCCAC
TTCACGATCAAAGATCCAGCTAACAGGCGCTACGAGGTGCCCTTGGAGACCCCGCGTGTC
CACAGCCGGGCACCGTCCCCACTCTACAGCGTGGAGTTCTCCGAGGAGCCCTTCGGGGTG
ATCGTGCACCGGCAGCTGGACGGCCGCGTGCTGCTGAACACGACGGTGGCGCCCCTGTTC
TTTGCGGACCAGTTCCTTCAGCTGTCCACCTCGCTGCCCTCGCAGTATATCACAGGCCTC
GCCGAGCACCTCAGTCCCCTGATGCTCAGCACCAGCTGGACCAGGATCACCCTGTGGAAC
CGGGACCTTGCGCCCACGCCCGGTGCGAACCTCTACGGGTCTCACCCTTTCTACCTGGCG
CTGGAGGACGGCGGGTCGGCACACGGGGTGTTCCTGCTAAACAGCAATGCCATGGATGTG
GTCCTGCAGCCGAGCCCTGCCCTTAGCTGGAGGTCGACAGGTGGGATCCTGGATGTCTAC
ATCTTCCTGGGCCCAGAGCCCAAGAGCGTGGTGCAGCAGTACCTGGACGTTGTGGGATAC
CCGTTCATGCCGCCATACTGGGGCCTGGGCTTCCACCTGTGCCGCTGGGGCTACTCCTCC
ACCGCTATCACCCGCCAGGTGGTGGAGAACATGACCAGGGCCCACTTCCCCCTGGACGTC
CAATGGAACGACCTGGACTACATGGACTCCCGGAGGGACTTCACGTTCAACAAGGATGGC
TTCCGGGACTTCCCGGCCATGGTGCAGGAGCTGCACCAGGGCGGCCGGCGCTACATGATG
ATCGTGGATCCTGCCATCAGCAGCTCGGGCCCTGCCGGGAGCTACAGGCCCTACGACGAG
GGTCTGCGGAGGGGGGTTTTCATCACCAACGAGACCGGCCAGCCGCTGATTGGGAAGGTA
TGGCCCGGGTCCACTGCCTTCCCCGACTTCACCAACCCCACAGCCCTGGCCTGGTGGGAG
GACATGGTGGCTGAGTTCCATGACCAGGTGCCCTTCGACGGCATGTGGATTGACATGAAC
GAGCCTTCCAACTTCATCAGAGGCTCTGAGGACGGCTGCCCCAACAATGAGCTGGAGAAC
CCACCCTACGTGCCTGGGGTGGTTGGGGGGACCCTCCAGGCGGCCACCATCTGTGCCTCC
AGCCACCAGTTTCTCTCCACACACTACAACCTGCACAACCTCTACGGCCTGACCGAAGCC
ATCGCCTCCCACAGGGCGCTGGTGAAGGCTCGGGGGACACGCCCATTTGTGATCTCCCGC
TCGACCTTTGCTGGCCACGGCCGATACGCCGGCCACTGGACGGGGGACGTGTGGAGCTCC
TGGGAGCAGCTCGCCTCCTCCGTGCCAGAAATCCTGCAGTTTAACCTGCTGGGGGTGCCT
CTGGTCGGGGCCGACGTCTGCGGCTTCCTGGGCAACACCTCAGAGGAGCTGTGTGTGCGC
TGGACCCAGCTGGGGGCCTTCTACCCCTTCATGCGGAACCACAACAGCCTGCTCAGTCTG
CCCCAGGAGCCGTACAGCTTCAGCGAGCCGGCCCAGCAGGCCATGAGGAAGGCCCTCACC
CTGCGCTACGCACTCCTCCCCCACCTCTACACACTGTTCCACCAGGCCCACGTCGCGGGG
GAGACCGTGGCCCGGCCCCTCTTCCTGGAGTTCCCCAAGGACTCTAGCACCTGGACTGTG
GACCACCAGCTCCTGTGGGGGGAGGCCCTGCTCATCACCCCAGTGCTCCAGGCCGGGAAG
GCCGAAGTGACTGGCTACTTCCCCTTGGGCACATGGTACGACCTGCAGACGGTGCCAATA
GAGGCCCTTGGCAGCCTCCCACCCCCACCTGCAGCTCCCCGTGAGCCAGCCATCCACAGC
GAGGGGCAGTGGGTGACGCTGCCGGCCCCCCTGGACACCATCAACGTCCACCTCCGGGCT
GGGTACATCATCCCCCTGCAGGGCCCTGGCCTCACAACCACAGAGTCCCGCCAGCAGCCC
ATGGCCCTGGCTGTGGCCCTGACCAAGGGTGGAGAGGCCCGAGGGGAGCTGTTCTGGGAC
GATGGAGAGAGCCTGGAAGTGCTGGAGCGAGGGGCCTACACACAGGTCATCTTCCTGGCC
AGGAATAACACGATCGTGAATGAGCTGGTACGTGTGACCAGTGAGGGAGCTGGCCTGCAG
CTGCAGAAGGTGACTGTCCTGGGCGTGGCCACGGCGCCCCAGCAGGTCCTCTCCAACGGT
GTCCCTGTCTCCAACTTCACCTACAGCCCCGACACCAAGGTCCTGGACATCTGTGTCTCG
CTGTTGATGGGAGAGCAGTTTCTCGTCAGCTGGTGTTAG
|
| Protein Properties |
|---|
| Number of Residues
| 952 |
| Molecular Weight
| Not Available |
| Theoretical pI
| Not Available |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Lysosomal alpha-glucosidase
MGVRHPPCSHRLLAVCALVSLATAALLGHILLHDFLLVPRELSGSSPVLEETHPAHQQGA
SRPGPRDAQAHPGRPRAVPTQCDVPPNSRFDCAPDKAITQEQCEARGCCYIPAKQGLQGA
QMGQPWCFFPPSYPSYKLENLSSSEMGYTATLTRTTPTFFPKDILTLRLDVMMETENRLH
FTIKDPANRRYEVPLETPHVHSRAPSPLYSVEFSEEPFGVIVHRQLDGRVLLNTTVAPLF
FADQFLQLSTSLPSQYITGLAEHLSPLMLSTSWTRITLWNRDLAPTPGANLYGSHPFYLA
LEDGGSAHGVFLLNSNAMDVVLQPSPALSWRSTGGILDVYIFLGPEPKSVVQQYLDVVGY
PFMPPYWGLGFHLCRWGYSSTAITRQVVENMTRAHFPLDVQWNDLDYMDSRRDFTFNKDG
FRDFPAMVQELHQGGRRYMMIVDPAISSSGPAGSYRPYDEGLRRGVFITNETGQPLIGKV
WPGSTAFPDFTNPTALAWWEDMVAEFHDQVPFDGMWIDMNEPSNFIRGSEDGCPNNELEN
PPYVPGVVGGTLQAATICASSHQFLSTHYNLHNLYGLTEAIASHRALVKARGTRPFVISR
STFAGHGRYAGHWTGDVWSSWEQLASSVPEILQFNLLGVPLVGADVCGFLGNTSEELCVR
WTQLGAFYPFMRNHNSLLSLPQEPYSFSEPAQQAMRKALTLRYALLPHLYTLFHQAHVAG
ETVARPLFLEFPKDSSTWTVDHQLLWGEALLITPVLQAGKAEVTGYFPLGTWYDLQTVPI
EALGSLPPPPAAPREPAIHSEGQWVTLPAPLDTINVHLRAGYIIPLQGPGLTTTESRQQP
MALAVALTKGGEARGELFWDDGESLEVLERGAYTQVIFLARNNTIVNELVRVTSEGAGLQ
LQKVTVLGVATAPQQVLSNGVPVSNFTYSPDTKVLDICVSLLMGEQFLVSWC
|
| External Links |
|---|
| GenBank ID Protein
| 31608 |
| UniProtKB/Swiss-Prot ID
| P10253 |
| UniProtKB/Swiss-Prot Entry Name
| LYAG_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| Y00839 |
| GeneCard ID
| GAA |
| GenAtlas ID
| GAA |
| HGNC ID
| HGNC:4065 |
| References |
|---|
| General References
| Not Available |

