| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-18 23:21:38 UTC |
|---|
| Updated at | 2020-12-07 19:06:55 UTC |
|---|
| CannabisDB ID | CDB006145 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Methylamine |
|---|
| Description | Methylamine, also known as aminomethane or methanamine, belongs to the class of organic compounds known as monoalkylamines. These are organic compounds containing a primary aliphatic amine group. Methylamine is a strong basic compound (based on its pKa). It has a strong odor similar to fish. Industrially, methylamine is used as a building block for the synthesis of many other commercially available compounds. Methylamine exists in all living organisms, ranging from bacteria to humans. Methylamine is found in relatively high concentrations in a number of plant foods such as wild carrots, carrots, and tea and in a lower concentration in cabbages, corns, and barley. Methylamine has also been detected, but not quantified in, several other plant foods, such as soybeans, wild celeries, french plantains, and common grapes. It has also been detected in cow’s milk. This could make methylamine a potential biomarker for the consumption of these foods. Methylamine occurs endogenously from amine catabolism and its tissue levels increase in some pathological conditions, including diabetes. In the human body methylamine and ammonia levels are reciprocally controlled by a semicarbazide-sensitive amine oxidase activity that deaminates methylamine to formaldehyde with the production of ammonia and hydrogen peroxide ( PMID: 18845121 ). Methylamine also targets the voltage-operated neuronal potassium channels, probably inducing release of neurotransmitter(s) ( PMID: 16049393 ). Semicarbazide-sensitive amine oxidase (SSAO) catalyzes the deamination of primary amines. This kind of deamination has been shown capable of regulating glucose transport in adipose cells. Increased serum SSAO activities have been found in patients with diabetic mellitus, vascular disorders, and Alzheimer's disease (PMID: 18691041 ). The SSAO-catalyzed deamination of endogenous substrates like methylamine is known to lead to the production of toxic formaldehyde. Chronic elevated methylamine increases the excretion of malondialdehyde and microalbuminuria. Amine oxidase substrates such as methylamine have been shown to stimulate glucose uptake by increasing the recruitment of the glucose transporter GLUT4 from vesicles within the cell to the cell surface. Methylamine has been identified as a uremic toxin according to the European Uremic Toxin Working Group (PMID: 22626821 ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| Aminomethane | ChEBI | | CH3-NH2 | ChEBI | | MeNH2 | ChEBI | | Methanamine | ChEBI | | MMA | ChEBI | | Monomethylamine | ChEBI | | Anhydrous methylamine | HMDB | | Carbinamine | HMDB | | Imizin | HMDB | | Mercurialin | HMDB | | Methyl group | HMDB | | Methyl OF gamma-N-methylasparagine | HMDB | | Methylamine anhydrous | HMDB | | Methylamine aqueous solution | HMDB | | Methylamine solution | HMDB | | Methylamine solutions | HMDB | | Methylaminen | HMDB | | Metilamine | HMDB | | Metyloamina | HMDB | | N-Methylamine | HMDB | | NMA | HMDB | | NME | HMDB | | Methylamine ion (1-) | HMDB | | Methylamine nitrate | HMDB | | Methylamine perchlorate | HMDB | | Methylamine sulfate (1:1) | HMDB | | Methylamine sulfate (2:1) | HMDB | | Methylamine, 13C-labeled | HMDB | | Methylamine, 14C-labeled | HMDB | | Methylamine, 15N-labeled | HMDB | | Methylamine hydrobromide | HMDB | | Methylamine hydrochloride, 14C-labeled | HMDB | | Methylamine, cesium salt | HMDB | | Methylamine, monopotassium salt | HMDB | | Methylammonium | HMDB | | Methylammonium ion | HMDB | | Methylamine hydroiodide | HMDB | | Methylamine, monosodium salt | HMDB | | Methylamine bisulfite | HMDB | | Methylamine hydride | HMDB | | Methylamine hydrochloride | HMDB | | Methylamine hydrofluoride | HMDB | | Methylamine hydrogen cyanide | HMDB | | Monomethylammonium ion | HMDB |
|
|---|
| Chemical Formula | CH5N |
|---|
| Average Molecular Weight | 31.06 |
|---|
| Monoisotopic Molecular Weight | 31.0422 |
|---|
| IUPAC Name | methanamine |
|---|
| Traditional Name | methylamine |
|---|
| CAS Registry Number | 74-89-5 |
|---|
| SMILES | CN |
|---|
| InChI Identifier | InChI=1S/CH5N/c1-2/h2H2,1H3 |
|---|
| InChI Key | BAVYZALUXZFZLV-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as monoalkylamines. These are organic compounds containing an primary aliphatic amine group. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic nitrogen compounds |
|---|
| Class | Organonitrogen compounds |
|---|
| Sub Class | Amines |
|---|
| Direct Parent | Monoalkylamines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Organopnictogen compound
- Hydrocarbon derivative
- Primary aliphatic amine
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Indirect biological role: Industrial application: Environmental role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | -93.4 °C | Not Available | | Boiling Point | −6.0 °C | Wikipedia | | Water Solubility | 1080 mg/mL at 25 °C | Not Available | | logP | -0.57 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-001i-9000000000-43b9d7b881c659f2ceb1 | 2014-09-20 | View Spectrum | | Predicted GC-MS | Methylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-001i-9000000000-50fb665f1ba89a03baf9 | Spectrum | | Predicted GC-MS | Methylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Methylamine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-001i-9000000000-93f5049fa2e67d0da26d | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-53c64a174764bada8913 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-001i-9000000000-1c10568342e547416eaf | 2012-07-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-2910620cfb01718df17f | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-2910620cfb01718df17f | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-2910620cfb01718df17f | 2017-07-25 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-9000000000-f140127f4f4b677d2975 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9000000000-f140127f4f4b677d2975 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-f140127f4f4b677d2975 | 2017-07-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-001i-9000000000-986b844d44ea8cab8877 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-001i-9000000000-986b844d44ea8cab8877 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001i-9000000000-986b844d44ea8cab8877 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-001i-9000000000-d90d418cf76ae365e6c5 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-001i-9000000000-d90d418cf76ae365e6c5 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-d90d418cf76ae365e6c5 | 2021-09-22 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 400 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Tyrosine Metabolism | 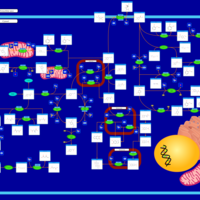 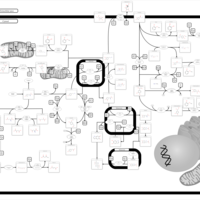 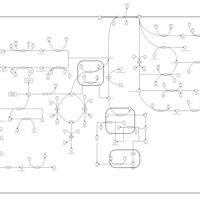 |  | | Alkaptonuria | 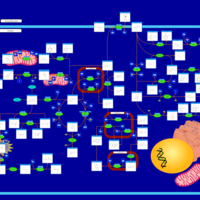 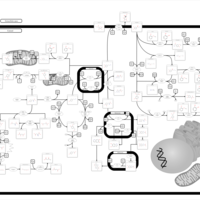 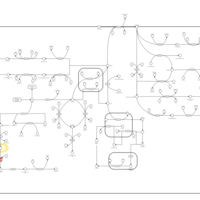 | Not Available | | Hawkinsinuria | 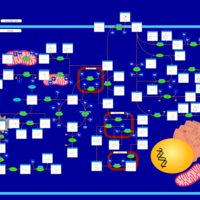  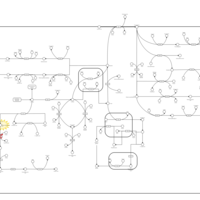 | Not Available | | Tyrosinemia Type I | 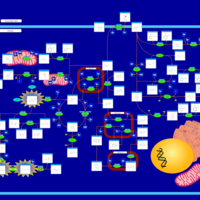 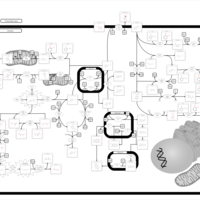 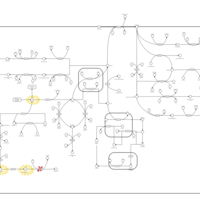 | Not Available | | Tyrosinemia, transient, of the newborn | 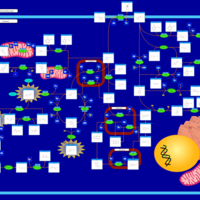 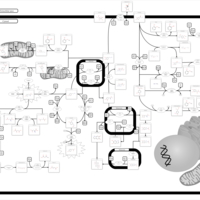 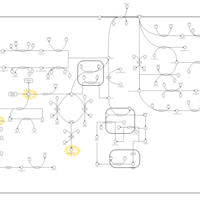 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | |
| Solute carrier family 2, facilitated glucose transporter member 4 | SLC2A4 | 17p13 | P14672 | details |
|
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000164 |
|---|
| DrugBank ID | DB01828 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB003958 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 6089 |
|---|
| KEGG Compound ID | C00218 |
|---|
| BioCyc ID | METHYLAMINE |
|---|
| BiGG ID | 1800349 |
|---|
| Wikipedia Link | Methylamine |
|---|
| METLIN ID | 3767 |
|---|
| PubChem Compound | 6329 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16830 |
|---|
| References |
|---|
| General References | - Xiao S, Yu PH: A fluorometric high-performance liquid chromatography procedure for simultaneous determination of methylamine and aminoacetone in blood and tissues. Anal Biochem. 2009 Jan 1;384(1):20-6. doi: 10.1016/j.ab.2008.09.029. Epub 2008 Sep 25. [PubMed:18845121 ]
- Pirisino R, Ghelardini C, De Siena G, Malmberg P, Galeotti N, Cioni L, Banchelli G, Raimondi L: Methylamine: a new endogenous modulator of neuron firing? Med Sci Monit. 2005 Aug;11(8):RA257-61. Epub 2005 Jul 25. [PubMed:16049393 ]
- Dunkel P, Gelain A, Barlocco D, Haider N, Gyires K, Sperlagh B, Magyar K, Maccioni E, Fadda A, Matyus P: Semicarbazide-sensitive amine oxidase/vascular adhesion protein 1: recent developments concerning substrates and inhibitors of a promising therapeutic target. Curr Med Chem. 2008;15(18):1827-39. doi: 10.2174/092986708785133022. [PubMed:18691041 ]
- Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, Vanholder R, Argiles A: Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012 Jul;23(7):1258-70. doi: 10.1681/ASN.2011121175. Epub 2012 May 24. [PubMed:22626821 ]
|
|---|
