| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-27 16:54:52 UTC |
|---|
| Updated at | 2021-01-04 18:49:15 UTC |
|---|
| CannabisDB ID | CDB005770 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Phenylacetic acid |
|---|
| Description | Phenylacetic acid, also known as phenylathanoic acid, phenylacetate, alpha tolylic acid or alpha-toluic acid, belongs to the class of organic compounds known as benzenes and substituted derivatives. These are aromatic compounds containing one monocyclic ring system consisting of benzene. Phenylacetic acid exists as a white solid that has a strong, honey-like odor. It is a catabolic product of phenylalanine. It is found in nearly all organisms from microbes to plants to animals. In plants it has been found to function as an auxin-like phyotohormone that is particularly enriched in fruits. However, it is not as strong as the basic auxin molecule, indole-3-acetic acid. Phenylacetate is used industrially in in some perfumes, as it possesses a honey-like odor even in low concentrations. Medically, phenylacetate is also employed to treat type II hyperammonemia to help reduce the amounts of ammonia in a patient's bloodstream by forming phenylacetyl-CoA, which then reacts with nitrogen-rich glutamine to form phenylacetylglutamine. This compound is then excreted from the patient's body. The sodium salt of phenylacetic acid, sodium phenylacetate, is used as a drug for the treatment of urea cycle disorders. Phenylacetic acid occurs naturally and has been detected, but not quantified in, several different foods, such as banana, fenugreeks, lemon thymes, pears, and peach (var.). This could make phenylacetic acid a potential biomarker for the consumption of these foods. Phenylacetic acid is also found in cannabis smoke and is formed during the combustion of cannabis ( Ref:DOI ). |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2-PHENYLACETIC ACID | ChEBI | | 2-Phenylethanoic acid | ChEBI | | alpha-Toluic acid | ChEBI | | Benzeneacetic acid | ChEBI | | Benzylformic acid | ChEBI | | Omega-phenylacetic acid | ChEBI | | PA | ChEBI | | Phenylacetate | Kegg | | 2-PHENYLACETate | Generator | | a-Toluate | Generator | | a-Toluic acid | Generator | | alpha-Toluate | Generator | | Α-toluate | Generator | | Α-toluic acid | Generator | | Benzeneacetate | Generator | | Benzylformate | Generator | | Omega-phenylacetate | Generator | | Phenylethanoate | HMDB | | W-Phenylacetate | HMDB | | W-Phenylacetic acid | HMDB | | Phenylacetic acid, potassium salt | HMDB | | Sodium phenylacetate | HMDB | | Phenylacetic acid, lithium salt | HMDB | | Phenylacetic acid, sodium salt | HMDB | | Phenylacetic acid, ammonium salt | HMDB | | Phenylacetic acid, calcium salt | HMDB | | Phenylacetic acid, cesium salt | HMDB | | Phenylacetic acid, mercury salt | HMDB | | Phenylacetic acid, rubidium salt | HMDB | | Phenylacetic acid, sodium salt , carboxy-(11)C-labeled CPD | HMDB | | PAA | PhytoBank | | ω-Phenylacetic acid | PhytoBank | | Phenylacetic acid | HMDB |
|
|---|
| Chemical Formula | C8H8O2 |
|---|
| Average Molecular Weight | 136.15 |
|---|
| Monoisotopic Molecular Weight | 136.0524 |
|---|
| IUPAC Name | 2-phenylacetic acid |
|---|
| Traditional Name | ω-phenylacetic acid |
|---|
| CAS Registry Number | 103-82-2 |
|---|
| SMILES | OC(=O)CC1=CC=CC=C1 |
|---|
| InChI Identifier | InChI=1S/C8H8O2/c9-8(10)6-7-4-2-1-3-5-7/h1-5H,6H2,(H,9,10) |
|---|
| InChI Key | WLJVXDMOQOGPHL-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as benzene and substituted derivatives. These are aromatic compounds containing one monocyclic ring system consisting of benzene. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Benzenoids |
|---|
| Class | Benzene and substituted derivatives |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Benzene and substituted derivatives |
|---|
| Alternative Parents | |
|---|
| Substituents | - Monocyclic benzene moiety
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework | Aromatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Biological location: Source: |
|---|
| Role | Indirect biological role: Industrial application: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 76.7 °C | Not Available | | Boiling Point | 265.5 °C | Wikipedia | | Water Solubility | 16.6 mg/mL | Not Available | | logP | 1.41 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0006-9100000000-1e23fd8e6ce900cd8e4c | 2014-09-20 | View Spectrum | | GC-MS | Phenylacetic acid, 1 TMS, GC-MS Spectrum | splash10-0006-9700000000-c07461cdad68959aa53f | Spectrum | | GC-MS | Phenylacetic acid, non-derivatized, GC-MS Spectrum | splash10-0006-9100000000-de65c7c0092343a4e599 | Spectrum | | GC-MS | Phenylacetic acid, non-derivatized, GC-MS Spectrum | splash10-0006-9700000000-c07461cdad68959aa53f | Spectrum | | GC-MS | Phenylacetic acid, non-derivatized, GC-MS Spectrum | splash10-0006-9700000000-43e86e45beae2fa6704f | Spectrum | | Predicted GC-MS | Phenylacetic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9200000000-dc43e5e062b0ed500c5f | Spectrum | | Predicted GC-MS | Phenylacetic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9100000000-aaef76d181aebe492bbe | Spectrum | | Predicted GC-MS | Phenylacetic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Phenylacetic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-0006-9100000000-de65c7c0092343a4e599 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-0006-9000000000-6ca33b098558bf13801c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-000i-0900000000-f898aafe4cecc87d26f1 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Positive | splash10-0006-9400000000-aa5bff1d7a8df7c97bdb | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-014i-9000000000-6a017c8136a375269f3e | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000i-2900000000-d79f9224f2390d6b17a2 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9000000000-e712b088c9a3d3855ec7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-01ri-9000000000-f0ed89dae2c22d4c538c | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-053i-2900000000-3e4bf20b985893f203e7 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-000i-1900000000-5419b00a9c7c126093db | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-000i-1900000000-9a2e089c5511fa2bf210 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kr-2900000000-94a6f40f937bf9d7e4c7 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9500000000-8d3540e40bfc0e4c0e5f | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0006-9000000000-417ad1f13a593ac780b9 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-000l-5900000000-0ed04a623daaf32fffda | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-000l-8900000000-709928d6b823240d3528 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9600000000-4a7739132344f3dec710 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0006-9000000000-5671d4c535a6553aaf0d | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0006-9000000000-5671d4c535a6553aaf0d | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-5e37e9f4d7b24c763302 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-00kf-9800000000-aacb25995ab02725e0f4 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0006-9100000000-930a2cbe3e9fb1915075 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-00kf-9000000000-199de8006c0d100aaabb | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 125 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 15.09 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Phenylacetate Metabolism | 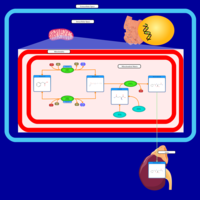 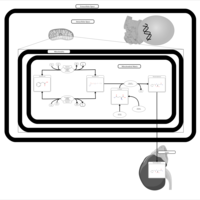  |  |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Aldehyde dehydrogenase, dimeric NADP-preferring | ALDH3A1 | 17p11.2 | P30838 | details | | Aldehyde dehydrogenase family 1 member A3 | ALDH1A3 | 15q26.3 | P47895 | details | | UDP-glucuronosyltransferase 1-1 | UGT1A1 | 2q37 | P22309 | details | | Aldehyde dehydrogenase family 3 member B2 | ALDH3B2 | 11q13 | P48448 | details | | Aldehyde dehydrogenase family 3 member B1 | ALDH3B1 | 11q13 | P43353 | details |
|
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | Not Available |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000209 |
|---|
| DrugBank ID | DB09269 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB031100 |
|---|
| KNApSAcK ID | C00000750 |
|---|
| Chemspider ID | 10181341 |
|---|
| KEGG Compound ID | C07086 |
|---|
| BioCyc ID | PHENYLACETATE |
|---|
| BiGG ID | 1486426 |
|---|
| Wikipedia Link | Phenylacetic_acid |
|---|
| METLIN ID | 129 |
|---|
| PubChem Compound | 999 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30745 |
|---|
| References |
|---|
| General References | Not Available |
|---|
