| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:22:22 UTC |
|---|
| Updated at | 2020-12-07 19:11:48 UTC |
|---|
| CannabisDB ID | CDB005230 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Xanthine |
|---|
| Description | Xanthine, also known as Xan or 2,6-dioxopurine, belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. An oxopurine in which the purine ring is substituted by oxo groups at positions 2 and 6 and N-9 is protonated. Xanthine is an extremely weak basic (essentially neutral) compound (based on its pKa). Xanthine exists in all living species, ranging from bacteria to humans. Within humans, xanthine participates in a number of enzymatic reactions. In particular, xanthine can be biosynthesized from guanine; which is mediated by the enzyme guanine deaminase. In addition, xanthine and ribose 1-phosphate can be biosynthesized from xanthosine through the action of the enzyme purine nucleoside phosphorylase. In humans, xanthine is involved in azathioprine action pathway. Xanthine is a potentially toxic compound. Xanthine, with regard to humans, has been found to be associated with several diseases such as hydrocephalus, adenosine kinase deficiency, xanthinuria type II, and eosinophilic esophagitis; xanthine has also been linked to the inborn metabolic disorder lesch-nyhan syndrome. Xanthine is expected to be in Cannabis as all living plants are known to produce and metabolize it. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 2,6-Dihydroxypurine | ChEBI | | 2,6-Dioxo-1,2,3,6-tetrahydropurine | ChEBI | | 9H-Purine-2,6-(1H,3H)-dione | ChEBI | | Purine-2(3H),6(1H)-dione | ChEBI | | Xan | ChEBI | | 1H-Purine-2,6-diol | HMDB | | 2,6(1,3)-Purinedion | HMDB | | 2,6-Dioxopurine | HMDB | | 3,7-Dihydro-1H-purine-2,6-dione | HMDB | | 3,7-Dihydropurine-2,6-dione | HMDB | | 9H-Purine-2,6(1H,3H)-dione | HMDB | | 9H-Purine-2,6-diol | HMDB | | Dioxopurine | HMDB | | Isoxanthine | HMDB | | Pseudoxanthine | HMDB | | Purine-2,6(1H,3H)-dione | HMDB | | Purine-2,6-diol | HMDB | | Xanthic oxide | HMDB | | Xanthin | HMDB |
|
|---|
| Chemical Formula | C5H4N4O2 |
|---|
| Average Molecular Weight | 152.11 |
|---|
| Monoisotopic Molecular Weight | 152.0334 |
|---|
| IUPAC Name | 2,3,6,7-tetrahydro-1H-purine-2,6-dione |
|---|
| Traditional Name | xanthine |
|---|
| CAS Registry Number | 69-89-6 |
|---|
| SMILES | O=C1NC2=C(NC=N2)C(=O)N1 |
|---|
| InChI Identifier | InChI=1S/C5H4N4O2/c10-4-2-3(7-1-6-2)8-5(11)9-4/h1H,(H3,6,7,8,9,10,11) |
|---|
| InChI Key | LRFVTYWOQMYALW-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as xanthines. These are purine derivatives with a ketone group conjugated at carbons 2 and 6 of the purine moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Imidazopyrimidines |
|---|
| Sub Class | Purines and purine derivatives |
|---|
| Direct Parent | Xanthines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Xanthine
- 6-oxopurine
- Purinone
- Alkaloid or derivatives
- Pyrimidone
- Pyrimidine
- Azole
- Imidazole
- Heteroaromatic compound
- Vinylogous amide
- Lactam
- Urea
- Azacycle
- Hydrocarbon derivative
- Organic oxide
- Organooxygen compound
- Organonitrogen compound
- Organic nitrogen compound
- Organopnictogen compound
- Organic oxygen compound
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework | Aromatic heteropolycyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | > 300 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 0.069 mg/mL at 16 °C; 9.5 mg/mL (sodium salt) | MERCK INDEX (1996); Human Metabolome Project (salt) | | logP | -0.73 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0udi-7900000000-2d5ab5d5db8ff4981467 | 2014-09-20 | View Spectrum | | GC-MS | Xanthine, non-derivatized, GC-MS Spectrum | splash10-0f6t-0924000000-9b80e0a2a60c73ca0180 | Spectrum | | GC-MS | Xanthine, non-derivatized, GC-MS Spectrum | splash10-0f6t-0924000000-9b80e0a2a60c73ca0180 | Spectrum | | GC-MS | Xanthine, non-derivatized, GC-MS Spectrum | splash10-0f6t-0924000000-30dc5892eecde860846a | Spectrum | | Predicted GC-MS | Xanthine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0kai-7900000000-2dc30b0fc4cff2239dbe | Spectrum | | Predicted GC-MS | Xanthine, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0udi-0900000000-a70539989d121bfacee0 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0a4i-6900000000-b047b06406308dbaeda8 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a4i-9300000000-ed480ed920c3e9b576ec | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0zfr-0900000000-efb049914c9bce596267 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF , negative | splash10-0zfr-0900000000-efb049914c9bce596267 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - , negative | splash10-0udi-0900000000-5fee91293851bb02193e | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , positive | splash10-000i-0900000000-4568a814903ff411923a | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9000000000-e078a358156f5a04d4c1 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-3900000000-250f7dc30d6fd96a4ef6 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0ik9-0900000000-3a6dad2473c1654789f6 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-a4e9443b51c3ac2fc58b | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Negative | splash10-00kf-9000000000-d23801a49488b1a5f0a2 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-4900000000-b177a2a36043e5e87efc | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0udi-0900000000-cb3b35c3117b7f36bf5c | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 10V, Negative | splash10-0zfr-0900000000-8d2a19e7c37d2cb7658f | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 30V, Negative | splash10-05mo-9300000000-33d00ab2288aaa055517 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 20V, Positive | splash10-0ik9-1900000000-3a6dad2473c1654789f6 | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 40V, Positive | splash10-001i-9000000000-37e76df2401a85967caa | 2021-09-20 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Negative | splash10-0udi-0900000000-4132da06dda895afc3ab | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0udi-0900000000-2e9e069e2df414aed037 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0w29-0900000000-fa52193346bc456d89e8 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a5i-9400000000-bbf70998e8b7515cb440 | 2015-05-26 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0udi-0900000000-566d663553ce4f0ec207 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0zfr-1900000000-d0a5d2c0f89f8d42d903 | 2015-05-27 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0006-9000000000-351d9f8ee3470f911829 | 2015-05-27 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100.7 MHz, DMSO-d6, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 1H]-TOCSY. Unexported temporarily by An Chi on Oct 15, 2021 until json or nmrML file is generated. 2D NMR Spectrum (experimental) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 100%_DMSO, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Purine Metabolism |  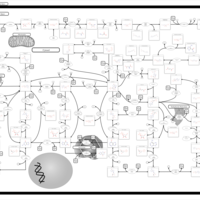 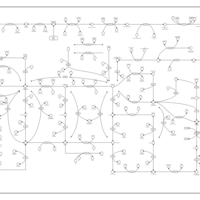 |  | | Adenosine Deaminase Deficiency |  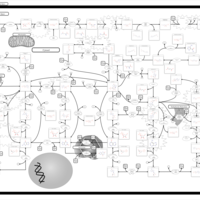 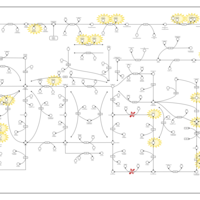 | Not Available | | Adenylosuccinate Lyase Deficiency | 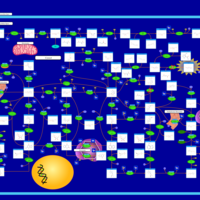 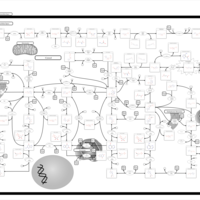 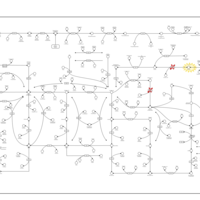 | Not Available | | Gout or Kelley-Seegmiller Syndrome | 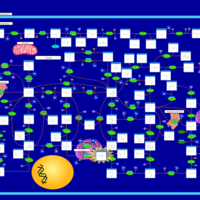 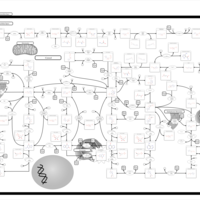 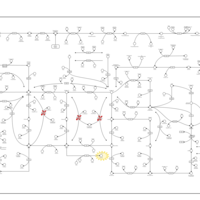 | Not Available | | Lesch-Nyhan Syndrome (LNS) | 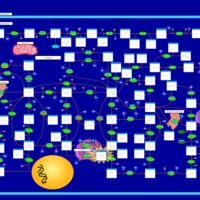 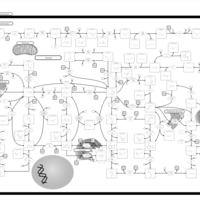  | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000292 |
|---|
| DrugBank ID | DB02134 |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB001977 |
|---|
| KNApSAcK ID | C00019660 |
|---|
| Chemspider ID | 1151 |
|---|
| KEGG Compound ID | C00385 |
|---|
| BioCyc ID | XANTHINE |
|---|
| BiGG ID | 34825 |
|---|
| Wikipedia Link | Xanthine |
|---|
| METLIN ID | 82 |
|---|
| PubChem Compound | 1188 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 17712 |
|---|
| References |
|---|
| General References | Not Available |
|---|
