| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-04-17 19:10:13 UTC |
|---|
| Updated at | 2021-01-04 18:59:20 UTC |
|---|
| CannabisDB ID | CDB005109 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Quinone |
|---|
| Description | Quinone also known as 1,4-benzoquinone, p-BQ or cyclohexadienedione, belongs to the class of organic compounds known as quinones. These are heterocyclic compounds containing two C=O bonds. Quinones are oxidized derivatives of aromatic compounds and are often readily made from reactive aromatic compounds with electron-donating substituents such as phenols and catechols, which increase the nucleophilicity of the ring and contributes to the large redox potential needed to break aromaticity. Quinone exists as a solid characterized by yellow prisms, with a penetrating odor resembling that of chlorine. It is used as a chemical intermediate, a polymerization inhibitor, an oxidizing agent, a photographic chemical, and a tanning agent. Occupational exposure to quinone may occur in the dye, textile, chemical, tanning, and cosmetic industries. Acute exposure to high levels of quinone, via inhalation in humans, is highly irritating to the eyes, resulting in discoloration of the conjunctiva and cornea. Chronic dermal contact to quinone in humans may result in skin ulceration, while chronic inhalation exposure may result in visual disturbances. Derivatives of quinones are common constituents of biologically relevant molecules. Some serve as electron acceptors in electron transport chains such as those in photosynthesis (plastoquinone, phylloquinone), and aerobic respiration (ubiquinone). Quinone is a common constituent of biologically relevant molecules (e.g. Vitamin K1 is phylloquinone). The auto-oxidation of the neurotransmitter dopamine and its precursor L-Dopa generates dopamine quinone which inhibits the functioning of dopamine transporter (DAT) and the TH enzyme leading to low mitochondrial ATP production (PMID: 24653659 ). Quinone is a constituent of tobacco and marijuana (cannabis) smoke and is formed during the combustion of cannabis ( Ref:DOI ). Quinone is quite toxic and is believed to be the causative agent for smoke-induced emphysema (PMID: 25057895 ). Rodent studies have shown that smoking causes progressive accumulation of quinone in the lung which leads to destruction of alveolar cells and emphysema. The pathogenesis involves arylation, oxidative stress, inflammation, and apoptosis (PMID: 25057895 ). The lung damage caused by smoke-generated quinone could be prevented by high doses of ascorbic acid (vitamin C), a known antagonist for quinone. |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1,4-Benzochinon | ChEBI | | 2,5-Cyclohexadiene-1,4-dione | ChEBI | | Benzo-1,4-quinone | ChEBI | | Benzoquinone | ChEBI | | p-Benzoquinone | ChEBI | | p-Chinon | ChEBI | | p-Quinone | ChEBI | | Para-benzoquinone | ChEBI | | Chinone | Kegg | | 1,4-Benzoquinone | Kegg | | 1,4-Benzoquine | HMDB | | 1,4-Cyclohexadiene dioxide | HMDB | | 1,4-Cyclohexadienedione | HMDB | | 1,4-Diossibenzene | HMDB | | 1,4-Dioxy-benzol | HMDB | | 1,4-Dioxybenzene | HMDB | | 2,5-Cyclohexadiene-1-4-dione | HMDB | | Benzo-chinon | HMDB | | Chinon | HMDB | | Cyclohexadiene-1,4-dione | HMDB | | Cyclohexadienedione | HMDB | | Eldoquin | HMDB | | Para-quinone | HMDB | | Quinone1,4-benzoquinone | HMDB | | Semiquinone anion | HMDB | | Semiquinone radicals | HMDB |
|
|---|
| Chemical Formula | C6H4O2 |
|---|
| Average Molecular Weight | 108.09 |
|---|
| Monoisotopic Molecular Weight | 108.0211 |
|---|
| IUPAC Name | cyclohexa-2,5-diene-1,4-dione |
|---|
| Traditional Name | quinone |
|---|
| CAS Registry Number | 106-51-4 |
|---|
| SMILES | O=C1C=CC(=O)C=C1 |
|---|
| InChI Identifier | InChI=1S/C6H4O2/c7-5-1-2-6(8)4-3-5/h1-4H |
|---|
| InChI Key | AZQWKYJCGOJGHM-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as p-benzoquinones. These are benzoquinones where the two C=O groups are attached at the 1- and 4-positions, respectively. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organic oxygen compounds |
|---|
| Class | Organooxygen compounds |
|---|
| Sub Class | Carbonyl compounds |
|---|
| Direct Parent | P-benzoquinones |
|---|
| Alternative Parents | |
|---|
| Substituents | - P-benzoquinone
- Organic oxide
- Hydrocarbon derivative
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework | Aliphatic homomonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Physiological effect | Health effect: |
|---|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Environmental role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | 115.7 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 11.1 mg/mL at 18 °C | Not Available | | logP | 0.20 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-0zgi-9200000000-8777cde0dbcbb2157cdd | 2014-09-20 | View Spectrum | | GC-MS | Quinone, non-derivatized, GC-MS Spectrum | splash10-0zgi-9500000000-c8bfbbc465fad7929f87 | Spectrum | | GC-MS | Quinone, non-derivatized, GC-MS Spectrum | splash10-00xr-1900000000-9b99afb0e9c6350434a4 | Spectrum | | GC-MS | Quinone, non-derivatized, GC-MS Spectrum | splash10-0zgi-9500000000-c8bfbbc465fad7929f87 | Spectrum | | GC-MS | Quinone, non-derivatized, GC-MS Spectrum | splash10-00xr-1900000000-9b99afb0e9c6350434a4 | Spectrum | | Predicted GC-MS | Quinone, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0a4i-9500000000-6664b553a87d4fc15f94 | Spectrum | | Predicted GC-MS | Quinone, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-0a4i-7900000000-fca2433f986ba378f8c9 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-00or-9000000000-6897ef153b4119e50ceb | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-014u-9000000000-9bddbff45c6d0d3e0cf1 | 2012-07-25 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (JEOL JMS-D-3000) , Positive | splash10-0zgi-9500000000-c8bfbbc465fad7929f87 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-QFT 7V, negative | splash10-014i-1900000000-6d13549d8c534563dc9b | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-QFT 9V, negative | splash10-014i-1900000000-af62dc0334e009fe3e5b | 2020-07-21 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - ESI-QFT 14V, negative | splash10-014i-3900000000-1167cf77177c0761168e | 2020-07-21 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0900000000-d886efcfed5b0135bcd5 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-1900000000-7f5c75e048ee2f454e33 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-9100000000-db1ffa9cf092dd5e4551 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-648d78080240a1294e4c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-0900000000-648d78080240a1294e4c | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4i-5900000000-0f04c7bee6397237ee21 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-0a4i-0900000000-2c6cb60a5040c6239920 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-0a4i-4900000000-f1f9c8ded0fe97bb684d | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-03di-9000000000-3c2c54b9382bddbdf0df | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0a4i-0900000000-62ea0dd9303aae554ccd | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-0a4i-2900000000-7966cc06756dd3b8c736 | 2021-09-24 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-004i-9000000000-28217a7272677bec662a | 2021-09-24 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, 5%_DMSO, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Riboflavin Metabolism | 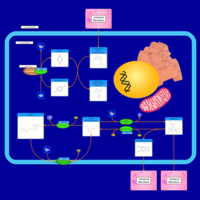 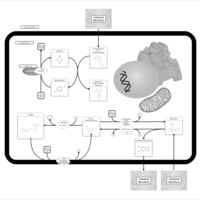 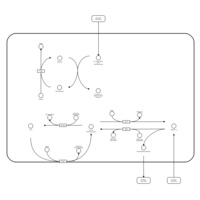 |  | | Pyrimidine Metabolism |  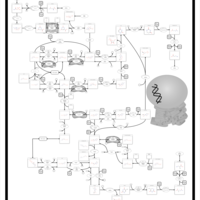  |  | | Beta Ureidopropionase Deficiency |  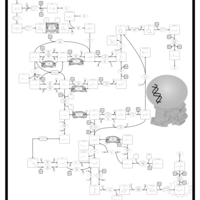 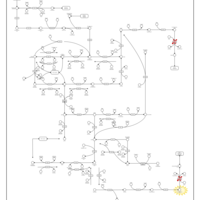 | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  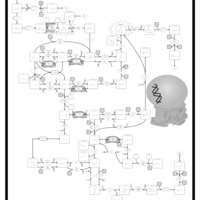 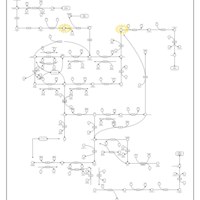 | Not Available | | Dihydropyrimidinase Deficiency |    | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
|---|
| Transporters | Not Available |
|---|
| Metal Bindings | |
|---|
| Receptors | |
| Amiloride-sensitive amine oxidase [copper-containing] | ABP1 | 7q36.1 | P19801 | details |
|
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0003364 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB005755 |
|---|
| KNApSAcK ID | Not Available |
|---|
| Chemspider ID | 4489 |
|---|
| KEGG Compound ID | C00472 |
|---|
| BioCyc ID | P-BENZOQUINONE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | 1,4-Benzoquinone |
|---|
| METLIN ID | 6905 |
|---|
| PubChem Compound | 4650 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 16509 |
|---|
| References |
|---|
| General References | - Dorszewska J, Prendecki M, Lianeri M, Kozubski W: Molecular Effects of L-dopa Therapy in Parkinson's Disease. Curr Genomics. 2014 Feb;15(1):11-7. doi: 10.2174/1389202914666131210213042. [PubMed:24653659 ]
- Ghosh A, Ganguly S, Dey N, Banerjee S, Das A, Chattopadhyay DJ, Chatterjee IB: Causation of cigarette smoke-induced emphysema by p-benzoquinone and its prevention by vitamin C. Am J Respir Cell Mol Biol. 2015 Mar;52(3):315-22. doi: 10.1165/rcmb.2013-0545OC. [PubMed:25057895 ]
|
|---|

