| Record Information |
|---|
| Version | 1.0 |
|---|
| Created at | 2020-03-19 00:34:00 UTC |
|---|
| Updated at | 2020-12-07 19:07:24 UTC |
|---|
| CannabisDB ID | CDB000380 |
|---|
| Secondary Accession Numbers | Not Available |
|---|
| Cannabis Compound Identification |
|---|
| Common Name | Caproic acid |
|---|
| Description | Caproic acid, also known as hexanoate or caproate, belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. Caproic acid is a medium chain triglyceride (MCT). Caproic acid is a medium chain triglycerides (MCT). Caproic acid is a very hydrophobic molecule, practically insoluble (in water), and relatively neutral. It is a conjugate acid of a hexanoate. Caproic acid is found naturally in various plant and animal fats and oils. Caproic acid is one of the saturated fatty acids that are known in cannabis plant. (PMID: 6991645 ) |
|---|
| Structure | |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Hexanoic acid | ChEBI | | 1-Pentanecarboxylic acid | ChEBI | | 6:0 | ChEBI | | Butylacetic acid | ChEBI | | C6:0 | ChEBI | | Capronic acid | ChEBI | | CH3-[CH2]4-COOH | ChEBI | | Hexanoate | ChEBI | | Hexoic acid | ChEBI | | Hexylic acid | ChEBI | | N-Caproic acid | ChEBI | | N-Hexanoic acid | ChEBI | | N-Hexoic acid | ChEBI | | N-Hexylic acid | ChEBI | | Pentanecarboxylic acid | ChEBI | | Pentiformic acid | ChEBI | | Pentylformic acid | ChEBI | | 1-Hexanoate | Generator | | 1-Pentanecarboxylate | Generator | | Butylacetate | Generator | | Capronate | Generator | | Hexanoic acid | Generator | | Hexoate | Generator | | Hexylate | Generator | | N-Caproate | Generator | | N-Hexanoate | Generator | | N-Hexoate | Generator | | N-Hexylate | Generator | | Pentanecarboxylate | Generator | | Pentiformate | Generator | | Pentylformate | Generator | | Caproate | Generator | | Hexanoic acid, calcium salt | MeSH, HMDB | | Hexanoic acid, sodium salt, 1-(11)C-labeled | MeSH, HMDB | | Hexanoic acid, nickel (2+) salt | MeSH, HMDB | | Hexanoic acid, sodium salt | MeSH, HMDB | | Bismuth(III)hexanoate | MeSH, HMDB | | Hexanoic acid, copper (2+) salt | MeSH, HMDB | | Hexanoic acid, manganese (2+) salt | MeSH, HMDB | | Bi(ohex)3 | MeSH, HMDB | | Hexanoic acid, barium salt | MeSH, HMDB | | Hexanoic acid, potassium salt | MeSH, HMDB | | Hexanoic acid, rhodium (2+) salt | MeSH, HMDB | | FA(6:0) | HMDB | | Calcium N-hexanoate | MeSH, HMDB | | Hexanoic acid, sodium salt (1:1) | MeSH, HMDB | | Sodium capronate | MeSH, HMDB | | Calcium hexanoate | MeSH, HMDB | | Caproic acid sodium salt | MeSH, HMDB | | Sodium caproate | MeSH, HMDB | | Sodium hexanoate | MeSH, HMDB |
|
|---|
| Chemical Formula | C6H12O2 |
|---|
| Average Molecular Weight | 116.16 |
|---|
| Monoisotopic Molecular Weight | 116.0837 |
|---|
| IUPAC Name | hexanoic acid |
|---|
| Traditional Name | hexanoic acid |
|---|
| CAS Registry Number | 142-62-1 |
|---|
| SMILES | CCCCCC(O)=O |
|---|
| InChI Identifier | InChI=1S/C6H12O2/c1-2-3-4-5-6(7)8/h2-5H2,1H3,(H,7,8) |
|---|
| InChI Key | FUZZWVXGSFPDMH-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as medium-chain fatty acids. These are fatty acids with an aliphatic tail that contains between 4 and 12 carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Fatty Acyls |
|---|
| Sub Class | Fatty acids and conjugates |
|---|
| Direct Parent | Medium-chain fatty acids |
|---|
| Alternative Parents | |
|---|
| Substituents | - Medium-chain fatty acid
- Straight chain fatty acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
|
| Disposition | Route of exposure: Source: Biological location: |
|---|
| Role | Industrial application: Biological role: |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Properties | | Property | Value | Reference |
|---|
| Melting Point | −3.4 °C | Wikipedia | | Boiling Point | 205.8 °C | Wikipedia | | Water Solubility | 10.3 mg/mL | Not Available | | logP | 1.92 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties | [] |
|---|
| Spectra |
|---|
| EI-MS/GC-MS | | Type | Description | Splash Key | View |
|---|
| EI-MS | Mass Spectrum (Electron Ionization) | splash10-03di-9000000000-fe3fab44832ddab1082e | 2014-09-20 | View Spectrum | | GC-MS | Caproic acid, 1 TMS, GC-MS Spectrum | splash10-00yi-3900000000-8d7942410dfae70c7c9d | Spectrum | | GC-MS | Caproic acid, non-derivatized, GC-MS Spectrum | splash10-03di-9000000000-f4bf00b677d90d54ba69 | Spectrum | | GC-MS | Caproic acid, non-derivatized, GC-MS Spectrum | splash10-00yi-3900000000-8d7942410dfae70c7c9d | Spectrum | | Predicted GC-MS | Caproic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | splash10-0006-9000000000-9ee618fa3c0bd9d56748 | Spectrum | | Predicted GC-MS | Caproic acid, 1 TMS, Predicted GC-MS Spectrum - 70eV, Positive | splash10-00fr-9200000000-be939d8c0455d1af93a6 | Spectrum | | Predicted GC-MS | Caproic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Caproic acid, non-derivatized, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum | | Predicted GC-MS | Caproic acid, TBDMS_1_1, Predicted GC-MS Spectrum - 70eV, Positive | Not Available | Spectrum |
|
|---|
| MS/MS | | Type | Description | Splash Key | View |
|---|
| MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-014i-0900000000-ba8c7078846c268d61a6 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-03di-0900000000-151d5971be58b6799d2e | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-02t9-1900000000-36096ef7dfebaa193910 | 2012-07-24 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - EI-B (HITACHI M-80B) , Positive | splash10-03di-9000000000-f4bf00b677d90d54ba69 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-014i-0900000000-f5b00c5e9d90d801ab16 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-014i-0900000000-5013166dd3b692762812 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-014i-5900000000-52b3e7f8b01498621772 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-e3af905666760ff49069 | 2012-08-31 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-014i-0900000000-f5b00c5e9d90d801ab16 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-014i-0900000000-5013166dd3b692762812 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-014i-5900000000-52b3e7f8b01498621772 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ , negative | splash10-0006-9000000000-e3af905666760ff49069 | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - LC-ESI-IT , negative | splash10-01b9-6900000000-763e9a8e0f15e13fae3c | 2017-09-14 | View Spectrum | | MS/MS | LC-MS/MS Spectrum - 35V, Positive | splash10-0002-9000000000-df670aaa002d77689cc7 | 2021-09-20 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-014j-9700000000-5489131065e0934751a7 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-05fs-9100000000-68f7c2883fb339578ace | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a4l-9000000000-8e5e925d60baa2f176c2 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-014i-2900000000-e877f22e42e8323f59e3 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01b9-9800000000-b5d583466919719819a1 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-0a4l-9000000000-791ecce51d7cdde8f0f5 | 2016-09-12 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-05tg-9000000000-b176d73a7b4fabde1ad4 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-006x-9000000000-08e066ae069da04a8689 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-052f-9000000000-808b7302aa744c7a1c20 | 2021-09-22 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-0002-9000000000-21c5ae8d52f2e0f4cd33 | 2021-09-23 | View Spectrum | | Predicted MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-004j-9000000000-3e67c250e6a67f63a0dd | 2021-09-23 | View Spectrum |
|
|---|
| NMR | | Type | Description | | View |
|---|
| 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 25.16 MHz, CDCl3, experimental) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, D2O, experimental) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | | Spectrum | | 2D NMR | [1H, 13C]-HSQC NMR Spectrum (2D, 600 MHz, H2O, experimental) | | Spectrum |
|
|---|
| Pathways |
|---|
| Pathways | | Name | SMPDB/Pathwhiz | KEGG | | Beta Oxidation of Very Long Chain Fatty Acids | 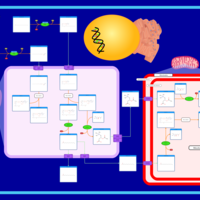 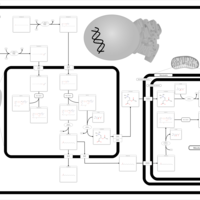 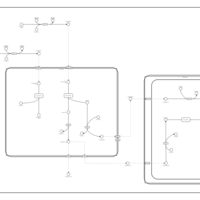 |  | | Fatty Acid Biosynthesis | 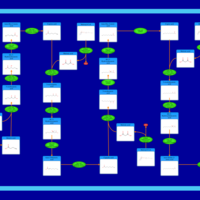 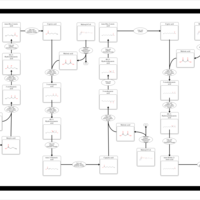 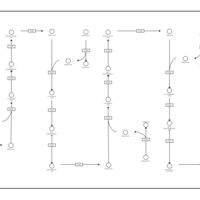 | Not Available | | Mitochondrial Beta-Oxidation of Short Chain Saturated Fatty Acids | 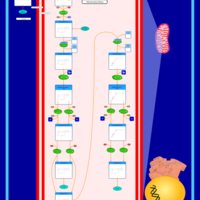 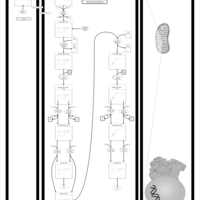  | Not Available | | Short-chain 3-hydroxyacyl-CoA dehydrogenase deficiency (SCHAD) | 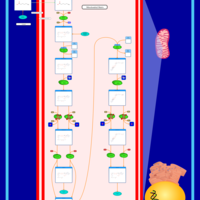 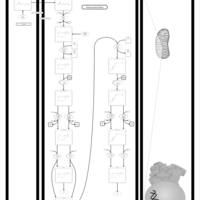 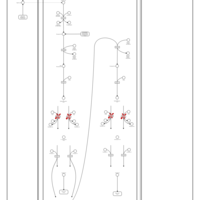 | Not Available |
|
|---|
| Protein Targets |
|---|
| Enzymes | |
| Peroxisomal carnitine O-octanoyltransferase | CROT | 7q21.1 | Q9UKG9 | details | | Carnitine O-palmitoyltransferase 2, mitochondrial | CPT2 | 1p32 | P23786 | details | | Mitochondrial carnitine/acylcarnitine carrier protein | SLC25A20 | 3p21.31 | O43772 | details | | Acyl-coenzyme A synthetase ACSM1, mitochondrial | ACSM1 | 16p12.3 | Q08AH1 | details | | Acyl-coenzyme A synthetase ACSM6, mitochondrial | ACSM6 | 10q23.33 | Q6P461 | details | | Acyl-coenzyme A synthetase ACSM2A, mitochondrial | ACSM2A | 16p12.3 | Q08AH3 | details | | Acyl-coenzyme A synthetase ACSM2B, mitochondrial | ACSM2B | 16p12.3 | Q68CK6 | details | | Acyl-coenzyme A synthetase ACSM3, mitochondrial | ACSM3 | 16p13.11 | Q53FZ2 | details | | Acyl-coenzyme A synthetase ACSM5, mitochondrial | ACSM5 | 16p12.3 | Q6NUN0 | details | | Acyl-coenzyme A synthetase ACSM4, mitochondrial | ACSM4 | 12p13.31 | P0C7M7 | details |
|
|---|
| Transporters | |
| Mitochondrial carnitine/acylcarnitine carrier protein | SLC25A20 | 3p21.31 | O43772 | details |
|
|---|
| Metal Bindings | |
| Acyl-coenzyme A synthetase ACSM1, mitochondrial | ACSM1 | 16p12.3 | Q08AH1 | details | | Acyl-coenzyme A synthetase ACSM6, mitochondrial | ACSM6 | 10q23.33 | Q6P461 | details | | Acyl-coenzyme A synthetase ACSM2A, mitochondrial | ACSM2A | 16p12.3 | Q08AH3 | details | | Acyl-coenzyme A synthetase ACSM2B, mitochondrial | ACSM2B | 16p12.3 | Q68CK6 | details | | Acyl-coenzyme A synthetase ACSM3, mitochondrial | ACSM3 | 16p13.11 | Q53FZ2 | details | | Acyl-coenzyme A synthetase ACSM5, mitochondrial | ACSM5 | 16p12.3 | Q6NUN0 | details | | Acyl-coenzyme A synthetase ACSM4, mitochondrial | ACSM4 | 12p13.31 | P0C7M7 | details |
|
|---|
| Receptors | Not Available |
|---|
| Transcriptional Factors | Not Available |
|---|
| Concentrations Data |
|---|
| Not Available |
|---|
| External Links |
|---|
| HMDB ID | HMDB0000535 |
|---|
| DrugBank ID | Not Available |
|---|
| Phenol Explorer Compound ID | Not Available |
|---|
| FoodDB ID | FDB013897 |
|---|
| KNApSAcK ID | C00001218 |
|---|
| Chemspider ID | 8552 |
|---|
| KEGG Compound ID | C01585 |
|---|
| BioCyc ID | HEXANOATE |
|---|
| BiGG ID | Not Available |
|---|
| Wikipedia Link | Hexanoic_acid |
|---|
| METLIN ID | 5520 |
|---|
| PubChem Compound | 8892 |
|---|
| PDB ID | Not Available |
|---|
| ChEBI ID | 30776 |
|---|
| References |
|---|
| General References | - Turner CE, Elsohly MA, Boeren EG: Constituents of Cannabis sativa L. XVII. A review of the natural constituents. J Nat Prod. 1980 Mar-Apr;43(2):169-234. doi: 10.1021/np50008a001. [PubMed:6991645 ]
|
|---|
