| Identification |
|---|
| HMDB Protein ID
| CDBP05240 |
| Secondary Accession Numbers
| Not Available |
| Name
| Probable ATP-dependent RNA helicase DDX58 |
| Description
| Not Available |
| Synonyms
|
- DEAD box protein 58
- RIG-I-like receptor 1
- Retinoic acid-inducible gene 1 protein
- Retinoic acid-inducible gene I protein
- RLR-1
- RIG-1
- RIG-I
|
| Gene Name
| DDX58 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Not Available |
| Specific Function
| Innate immune receptor which acts as a cytoplasmic sensor of viral nucleic acids and plays a major role in sensing viral infection and in the activation of a cascade of antiviral responses including the induction of type I interferons and proinflammatory cytokines. Its ligands include: 5'-triphosphorylated ssRNA and dsRNA and short dsRNA (<1 kb in length). In addition to the 5'-triphosphate moiety, blunt-end base pairing at the 5'-end of the RNA is very essential. Overhangs at the non-triphosphorylated end of the dsRNA RNA have no major impact on its activity. A 3'overhang at the 5'triphosphate end decreases and any 5'overhang at the 5' triphosphate end abolishes its activity. Upon ligand binding it associates with mitochondria antiviral signaling protein (MAVS/IPS1) which activates the IKK-related kinases: TBK1 and IKBKE which phosphorylate interferon regulatory factors: IRF3 and IRF7 which in turn activate transcription of antiviral immunological genes, including interferons (IFNs); IFN-alpha and IFN-beta. Detects both positive and negative strand RNA viruses including members of the families Paramyxoviridae: Human respiratory syncytial virus and measles virus (MeV), Rhabdoviridae: vesicular stomatitis virus (VSV), Orthomyxoviridae: influenza A and B virus, Flaviviridae: Japanese encephalitis virus (JEV), hepatitis C virus (HCV), dengue virus (DENV) and west Nile virus (WNV). It also detects rotavirus and reovirus. Also involved in antiviral signaling in response to viruses containing a dsDNA genome such as Epstein-Barr virus (EBV). Detects dsRNA produced from non-self dsDNA by RNA polymerase III, such as Epstein-Barr virus-encoded RNAs (EBERs). May play important roles in granulocyte production and differentiation, bacterial phagocytosis and in the regulation of cell migration.
|
| GO Classification
|
| Biological Process |
| positive regulation of defense response to virus by host |
| positive regulation of transcription from RNA polymerase II promoter |
| response to exogenous dsRNA |
| positive regulation of interferon-beta production |
| detection of virus |
| negative regulation of type I interferon production |
| positive regulation of interferon-alpha production |
| positive regulation of sequence-specific DNA binding transcription factor activity |
| positive regulation of transcription factor import into nucleus |
| regulation of cell migration |
| regulation of type III interferon production |
| RIG-I signaling pathway |
| innate immune response |
| virus-host interaction |
| Cellular Component |
| cytosol |
| actin cytoskeleton |
| ruffle membrane |
| tight junction |
| Molecular Function |
| metal ion binding |
| double-stranded RNA binding |
| ATP binding |
| double-stranded DNA binding |
| single-stranded RNA binding |
| ATP-dependent helicase activity |
| zinc ion binding |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | NF-kappa B signaling pathway | Not Available | 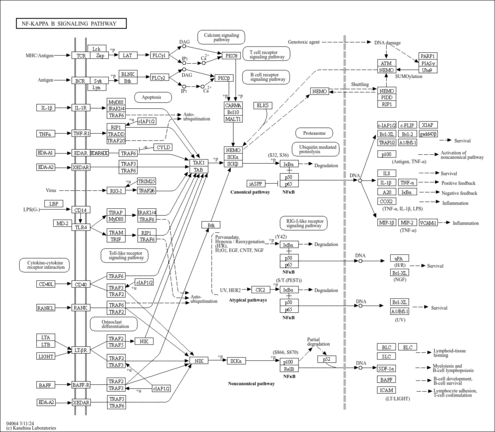 | | RIG-I-like receptor signaling pathway | Not Available | 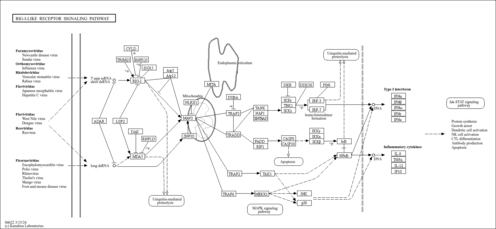 | | Cytosolic DNA-sensing pathway | Not Available | 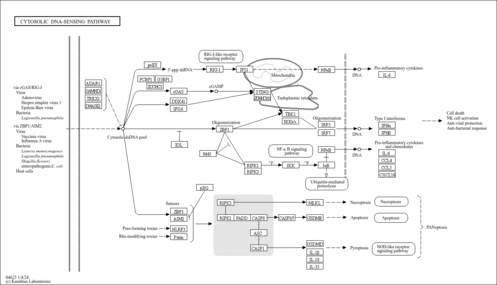 | | Hepatitis C | Not Available | 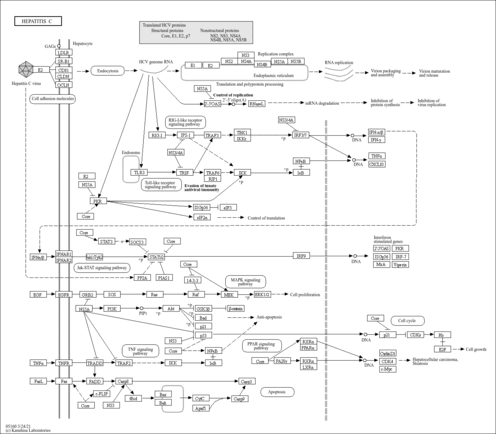 | | Hepatitis B | Not Available |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 9 |
| Locus
| 9p12 |
| SNPs
| Not Available |
| Gene Sequence
|
Not Available
|
| Protein Properties |
|---|
| Number of Residues
| Not Available |
| Molecular Weight
| 106598.72 |
| Theoretical pI
| 6.407 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>gi|27881482|ref|NP_055129.2| probable ATP-dependent RNA helicase DDX58 [Homo sapiens]
MTTEQRRSLQAFQDYIRKTLDPTYILSYMAPWFREEEVQYIQAEKNNKGPMEAATLFLKF
LLELQEEGWF
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| O95786 |
| UniProtKB/Swiss-Prot Entry Name
| Not Available |
| PDB IDs
|
|
| GenBank Gene ID
| Not Available |
| GeneCard ID
| Not Available |
| GenAtlas ID
| Not Available |
| HGNC ID
| HGNC:19102 |
| References |
|---|
| General References
| Not Available |




