| Identification |
|---|
| HMDB Protein ID
| CDBP04791 |
| Secondary Accession Numbers
| Not Available |
| Name
| DNA replication licensing factor MCM7 |
| Description
| Not Available |
| Synonyms
|
- CDC47 homolog
- P1.1-MCM3
|
| Gene Name
| MCM7 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in nucleotide binding |
| Specific Function
| Acts as component of the MCM2-7 complex (MCM complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the MCM2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. Required for S-phase checkpoint activation upon UV-induced damage.
|
| GO Classification
|
| Biological Process |
| response to DNA damage stimulus |
| cell cycle checkpoint |
| DNA strand elongation involved in DNA replication |
| DNA replication initiation |
| M/G1 transition of mitotic cell cycle |
| DNA unwinding involved in replication |
| regulation of phosphorylation |
| cell proliferation |
| S phase of mitotic cell cycle |
| G1/S transition of mitotic cell cycle |
| Cellular Component |
| nucleoplasm |
| chromatin |
| MCM complex |
| Component |
| organelle |
| membrane-bounded organelle |
| intracellular membrane-bounded organelle |
| nucleus |
| Function |
| catalytic activity |
| hydrolase activity |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| nucleic acid binding |
| dna binding |
| nucleoside-triphosphatase activity |
| hydrolase activity, acting on acid anhydrides |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| pyrophosphatase activity |
| binding |
| nucleotide binding |
| Molecular Function |
| ATP binding |
| single-stranded DNA binding |
| DNA helicase activity |
| Process |
| metabolic process |
| macromolecule metabolic process |
| cellular macromolecule metabolic process |
| dna metabolic process |
| dna replication |
| dna-dependent dna replication initiation |
|
| Cellular Location
|
- Nucleus
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | DNA replication | Not Available | 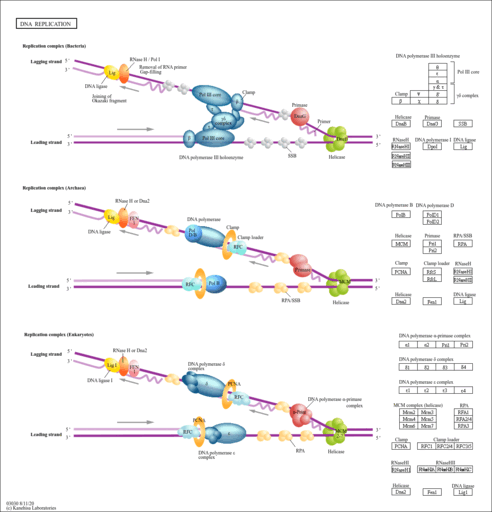 | | Cell cycle | Not Available | 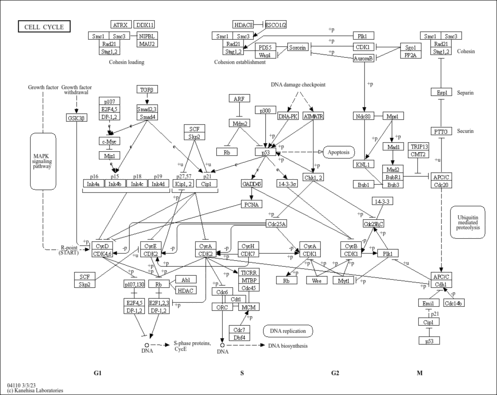 |
|
| Gene Properties |
|---|
| Chromosome Location
| 7 |
| Locus
| 7q21.3-q22.1 |
| SNPs
| MCM7 |
| Gene Sequence
|
>2160 bp
ATGGCACTGAAGGACTACGCGCTAGAGAAGGAAAAGGTTAAGAAGTTCTTACAAGAGTTC
TACCAGGATGATGAACTCGGGAAGAAGCAGTTCAAGTATGGGAACCAGTTGGTTCGGCTG
GCTCATCGGGAACAGGTGGCTCTGTATGTGGACCTGGACGACGTAGCCGAGGATGACCCC
GAGTTGGTGGACTCAATTTGTGAGAATGCCAGGCGCTACGCGAAGCTCTTTGCTGATGCC
GTACAAGAGCTGCTGCCTCAGTACAAGGAGAGGGAAGTGGTAAATAAAGATGTCCTGGAC
GTTTACATTGAGCATCGGCTAATGATGGAGCAGCGGAGTCGGGACCCTGGGATGGTCCGA
AGCCCCCAGAACCAGTACCCTGCTGAACTCATGCGCAGATTTGAGCTGTATTTTCAAGGC
CCTAGCAGCAACAAGCCTCGTGTGATCCGGGAAGTGCGGGCTGACTCTGTGGGGAAGTTG
GTAACTGTGCGTGGAATCGTCACTCGTGTCTCTGAAGTCAAACCCAAGATGGTGGTGGCC
ACTTACACTTGTGACCAGTGTGGGGCAGAGACCTACCAGCCGATCCAGTCTCCCACTTTC
ATGCCTCTGATCATGTGCCCAAGCCAGGAGTGCCAAACCAACCGCTCAGGAGGGCGGCTG
TATCTGCAGACACGGGGCTCCAGATTCATCAAATTCCAGGAGATGAAGATGCAAGAACAT
AGTGATCAGGTGCCTGTGGGAAATATCCCTCGTAGTATCACGGTGCTGGTAGAAGGAGAG
AACACAAGGATTGCCCAGCCTGGAGACCACGTCAGCGTCACTGGTATTTTCTTGCCAATC
CTGCGCACTGGGTTCCGACAGGTGGTACAGGGTTTACTCTCAGAAACCTACCTGGAAGCC
CATCGGATTGTGAAGATGAACAAGAGTGAGGATGATGAGTCTGGGGCTGGAGAGCTCACC
AGGGAGGAGCTGAGGCAAATTGCAGAGGAGGATTTCTACGAAAAGCTGGCAGCTTCAATC
GCCCCAGAAATATACGGGCATGAAGATGTGAAGAAGGCACTGCTGCTCCTGCTAGTCGGG
GGTGTGGACCAGTCTCCTCGAGGCATGAAAATCCGGGGCAACATCAACATCTGTCTGATG
GGGGATCCTGGTGTGGCCAAGTCTCAGCTCCTGTCATACATTGATCGACTGGCGCCTCGC
AGCCAGTACACAACAGGCCGGGGCTCCTCAGGAGTGGGGCTTACGGCAGCTGTGCTGAGA
GACTCCGTGAGTGGAGAACTGACCTTAGAGGGTGGGGCCCTGGTGCTGGCTGACCAGGGT
GTGTGCTGCATTGATGAGTTCGACAAGATGGCTGAGGCCGACCGCACAGCCATCCACGAG
GTCATGGAGCAGCAGACCATCTCCATTGCCAAGGCCGGCATTCTCACCACACTCAATGCC
CGCTGCTCCATCCTGGCTGCCGCCAACCCTGCCTACGGGCGCTACAACCCTCGCCGCAGC
CTGGAGCAGAACATACAGCTACCTGCTGCACTGCTCTCCCGGTTTGACCTCCTCTGGCTG
ATTCAGGACCGGCCCGACCGAGACAATGACCTACGGTTGGCCCAGCACATCACCTATGTG
CACCAGCACAGCCGGCAGCCCCCCTCCCAGTTTGAACCTCTGGACATGAAGCTCATGAGG
CGTTACATAGCCATGTGCCGCGAGAAGCAGCCCATGGTGCCAGAGTCTCTGGCTGACTAC
ATCACAGCAGCATACGTGGAGATGAGGCGAGAGGCTTGGGCTAGTAAGGATGCCACCTAT
ACTTCTGCCCGGACCCTGCTGGCTATCCTGCGCCTTTCCACTGCTCTGGCACGTCTGAGA
ATGGTGGATGTGGTGGAGAAAGAAGATGTGAATGAAGCCATCAGGCTAATGGAGATGTCA
AAGGACTCTCTTCTAGGAGACAAGGGGCAGACAGCTAGGACTCAGAGACCAGCAGATGTG
ATATTTGCCACCGTCCGTGAACTGGTCTCAGGGGGCCGAAGTGTCCGGTTCTCTGAGGCA
GAGCAGCGCTGTGTATCTCGTGGCTTCACACCCGCCCAGTTCCAGGCGGCTCTGGATGAA
TATGAGGAGCTCAATGTCTGGCAGGTCAATGCTTCCCGGACACGGATCACTTTTGTCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 719 |
| Molecular Weight
| 81307.22 |
| Theoretical pI
| 6.454 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>DNA replication licensing factor MCM7
MALKDYALEKEKVKKFLQEFYQDDELGKKQFKYGNQLVRLAHREQVALYVDLDDVAEDDP
ELVDSICENARRYAKLFADAVQELLPQYKEREVVNKDVLDVYIEHRLMMEQRSRDPGMVR
SPQNQYPAELMRRFELYFQGPSSNKPRVIREVRADSVGKLVTVRGIVTRVSEVKPKMVVA
TYTCDQCGAETYQPIQSPTFMPLIMCPSQECQTNRSGGRLYLQTRGSRFIKFQEMKMQEH
SDQVPVGNIPRSITVLVEGENTRIAQPGDHVSVTGIFLPILRTGFRQVVQGLLSETYLEA
HRIVKMNKSEDDESGAGELTREELRQIAEEDFYEKLAASIAPEIYGHEDVKKALLLLLVG
GVDQSPRGMKIRGNINICLMGDPGVAKSQLLSYIDRLAPRSQYTTGRGSSGVGLTAAVLR
DSVSGELTLEGGALVLADQGVCCIDEFDKMAEADRTAIHEVMEQQTISIAKAGILTTLNA
RCSILAAANPAYGRYNPRRSLEQNIQLPAALLSRFDLLWLIQDRPDRDNDLRLAQHITYV
HQHSRQPPSQFEPLDMKLMRRYIAMCREKQPMVPESLADYITAAYVEMRREAWASKDATY
TSARTLLAILRLSTALARLRMVDVVEKEDVNEAIRLMEMSKDSLLGDKGQTARTQRPADV
IFATVRELVSGGRSVRFSEAEQRCVSRGFTPAQFQAALDEYEELNVWQVNASRTRITFV
|
| External Links |
|---|
| GenBank ID Protein
| 33469968 |
| UniProtKB/Swiss-Prot ID
| P33993 |
| UniProtKB/Swiss-Prot Entry Name
| MCM7_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| NM_005916.3 |
| GeneCard ID
| MCM7 |
| GenAtlas ID
| MCM7 |
| HGNC ID
| HGNC:6950 |
| References |
|---|
| General References
| Not Available |

