| Identification |
|---|
| HMDB Protein ID
| CDBP04667 |
| Secondary Accession Numbers
| Not Available |
| Name
| Spliceosome RNA helicase DDX39B |
| Description
| Not Available |
| Synonyms
|
- 56 kDa U2AF65-associated protein
- ATP-dependent RNA helicase p47
- DEAD box protein UAP56
- HLA-B-associated transcript 1 protein
|
| Gene Name
| DDX39B |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in nucleic acid binding |
| Specific Function
| Component of the THO subcomplex of the TREX complex. The TREX complex specifically associates with spliced mRNA and not with unspliced pre-mRNA. It is recruited to spliced mRNAs by a transcription-independent mechanism. Binds to mRNA upstream of the exon-junction complex (EJC) and is recruited in a splicing- and cap-dependent manner to a region near the 5' end of the mRNA where it functions in mRNA export. The recruitment occurs via an interaction between ALYREF/THOC4 and the cap-binding protein NCBP1. DDX39B functions as a bridge between ALYREF/THOC4 and the THO complex. The TREX complex is essential for the export of Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs and infectious virus production. The recruitment of the TREX complex to the intronless viral mRNA occurs via an interaction between KSHV ORF57 protein and ALYREF/THOC4.
Splice factor that is required for the first ATP-dependent step in spliceosome assembly and for the interaction of U2 snRNP with the branchpoint. Has both RNA-stimulated ATP binding/hydrolysis activity and ATP-dependent RNA unwinding activity. Even with the stimulation of RNA, the ATPase activity is weak. Can only hydrolyze ATP but not other NTPs. The RNA stimulation of ATPase activity does not have a strong preference for the sequence and length of the RNA. However, ssRNA stimulates the ATPase activity much more strongly than dsRNA. Can unwind 5' or 3' overhangs or blunt end RNA duplexes in vitro. The ATPase and helicase activities are not influenced by U2AF2 and ALYREF/THOC4.
|
| GO Classification
|
| Biological Process |
| spliceosomal complex assembly |
| RNA secondary structure unwinding |
| intronless viral mRNA export from host nucleus |
| Cellular Component |
| cytoplasm |
| spliceosomal complex |
| nuclear speck |
| transcription export complex |
| Function |
| atpase activity |
| atpase activity, coupled |
| nucleic acid binding |
| nucleoside-triphosphatase activity |
| helicase activity |
| hydrolase activity, acting on acid anhydrides |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| pyrophosphatase activity |
| binding |
| catalytic activity |
| hydrolase activity |
| nucleoside binding |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| atp-dependent helicase activity |
| Molecular Function |
| ATP-dependent RNA helicase activity |
| ATP binding |
| ATP-dependent protein binding |
| U4 snRNA binding |
| U6 snRNA binding |
|
| Cellular Location
|
- Nucleus
- Nucleus speckle
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | RNA transport | Not Available | 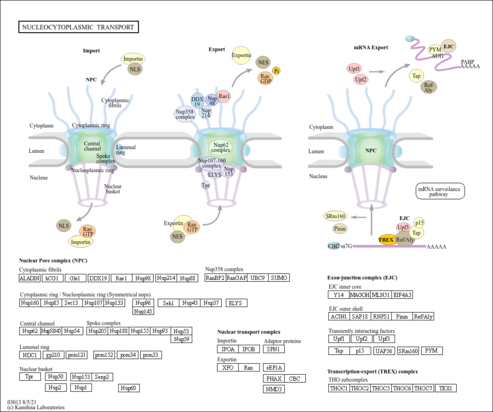 | | mRNA surveillance pathway | Not Available | 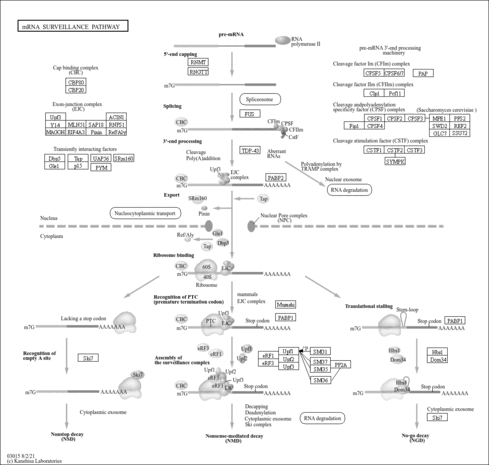 | | Spliceosome | Not Available | 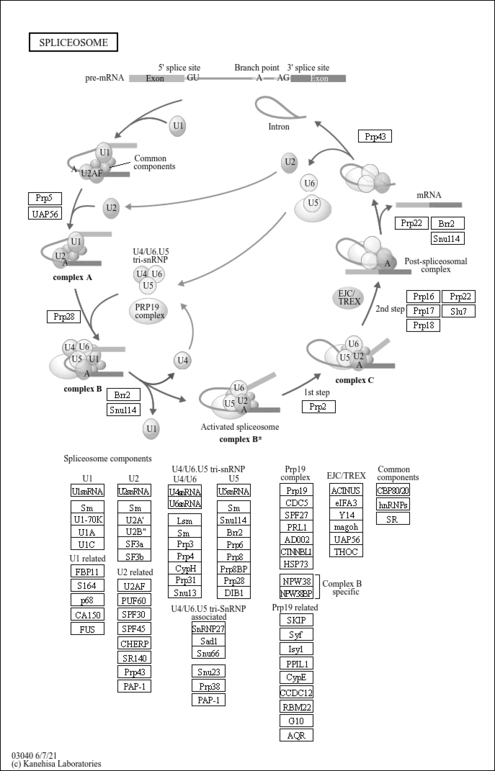 | | Influenza A | Not Available | 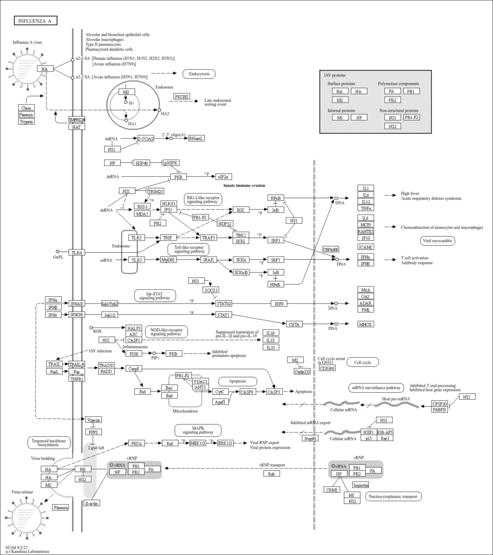 |
|
| Gene Properties |
|---|
| Chromosome Location
| 6 |
| Locus
| 6p21.3 |
| SNPs
| BAT1 |
| Gene Sequence
|
>1287 bp
ATGGCAGAGAACGATGTGGACAATGAGCTCTTGGACTATGAAGATGATGAGGTGGAGACA
GCAGCTGGGGGAGATGGGGCTGAGGCCCCTGCCAAGAAGGATGTCAAGGGCTCCTATGTC
TCCATCCACAGCTCTGGCTTTCGTGACTTCCTGCTCAAGCCAGAGTTGCTCCGGGCCATT
GTCGACTGTGGCTTTGAGCATCCGTCAGAAGTCCAGCATGAGTGCATCCCTCAGGCCATT
CTGGGAATGGATGTCCTGTGCCAGGCCAAGTCGGGCATGGGAAAGACAGCAGTGTTTGTC
TTGGCCACACTGCAACAGCTGGAGCCAGTTACTGGGCAGGTGTCTGTACTGGTGATGTGT
CACACTCGGGAGTTGGCTTTTCAGATCAGCAAGGAATATGAGCGCTTCTCTAAATACATG
CCCAATGTCAAGGTTGCTGTTTTTTTTGGTGGTCTGTCTATCAAGAAGGATGAAGAGGTG
CTGAAGAAGAACTGCCCGCATATCGTCGTGGGGACTCCAGGCCGTATCCTAGCCCTGGCT
CGAAATAAGAGCCTCAACCTCAAACACATTAAACACTTTATTTTGGATGAATGTGATAAG
ATGCTTGAACAGCTCGACATGCGTCGGGATGTCCAGGAAATTTTTCGCATGACCCCCCAC
GAGAAGCAGGTCATGATGTTCAGTGCTACCTTGAGCAAAGAGATCCGTCCAGTCTGCCGC
AAGTTCATGCAAGATCCAATGGAGATCTTCGTGGATGATGAGACGAAGTTGACGCTGCAT
GGGTTGCAGCAGTACTACGTGAAACTGAAGGACAACGAGAAGAACCGGAAGCTCTTTGAC
CTTCTGGATGTCCTTGAGTTCAACCAGGTGGTGATCTTTGTGAAGTCTGTGCAGCGGTGC
ATTGCCTTGGCCCAGCTACTAGTGGAGCAGAACTTCCCAGCCATTGCCATCCACCGTGGG
ATGCCCCAGGAGGAGAGGCTTTCTCGGTATCAGCAGTTTAAAGATTTTCAACGACGAATT
CTTGTGGCTACCAACCTATTTGGCCGAGGCATGGACATCGAGCGGGTGAACATTGCTTTT
AATTATGACATGCCTGAGGATTCTGACACCTACCTGCATCGGGTGGCCAGAGCAGGCCGG
TTTGGCACCAAGGGCTTGGCTATCACATTTGTGTCCGATGAGAATGATGCCAAGATCCTC
AATGATGTGCAGGATCGCTTTGAGGTCAATATTAGTGAGCTGCCTGATGAGATAGACATC
TCCTCCTACATTGAACAGACACGGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 428 |
| Molecular Weight
| 48990.945 |
| Theoretical pI
| 5.668 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Spliceosome RNA helicase BAT1
MAENDVDNELLDYEDDEVETAAGGDGAEAPAKKDVKGSYVSIHSSGFRDFLLKPELLRAI
VDCGFEHPSEVQHECIPQAILGMDVLCQAKSGMGKTAVFVLATLQQLEPVTGQVSVLVMC
HTRELAFQISKEYERFSKYMPNVKVAVFFGGLSIKKDEEVLKKNCPHIVVGTPGRILALA
RNKSLNLKHIKHFILDECDKMLEQLDMRRDVQEIFRMTPHEKQVMMFSATLSKEIRPVCR
KFMQDPMEIFVDDETKLTLHGLQQYYVKLKDNEKNRKLFDLLDVLEFNQVVIFVKSVQRC
IALAQLLVEQNFPAIAIHRGMPQEERLSRYQQFKDFQRRILVATNLFGRGMDIERVNIAF
NYDMPEDSDTYLHRVARAGRFGTKGLAITFVSDENDAKILNDVQDRFEVNISELPDEIDI
SSYIEQTR
|
| External Links |
|---|
| GenBank ID Protein
| 587146 |
| UniProtKB/Swiss-Prot ID
| Q13838 |
| UniProtKB/Swiss-Prot Entry Name
| UAP56_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| Z37166 |
| GeneCard ID
| BAT1 |
| GenAtlas ID
| BAT1 |
| HGNC ID
| HGNC:13917 |
| References |
|---|
| General References
| Not Available |



