| Identification |
|---|
| HMDB Protein ID
| CDBP04663 |
| Secondary Accession Numbers
| Not Available |
| Name
| Alkaline ceramidase 1 |
| Description
| Not Available |
| Synonyms
|
- Acylsphingosine deacylase 3
- AlkCDase 1
- Alkaline CDase 1
- N-acylsphingosine amidohydrolase 3
|
| Gene Name
| ACER1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides |
| Specific Function
| Hydrolyzes the sphingolipid ceramide into sphingosine and free fatty acid at an optimal pH of 8.0. Has a highly restricted substrate specificity for the natural stereoisomer of ceramide with D-erythro-sphingosine but not D-ribo-phytosphingosine or D-erythro-dihydrosphingosine as a backbone. May have a role in regulating the levels of bioactive lipids ceramide and sphingosine 1-phosphate, as well as complex sphingolipids (By similarity).
|
| GO Classification
|
| Biological Process |
| keratinocyte differentiation |
| phospholipid metabolic process |
| regulation of lipid metabolic process |
| response to alkalinity |
| sphingosine biosynthetic process |
| cellular response to calcium ion |
| ceramide catabolic process |
| Cellular Component |
| endoplasmic reticulum membrane |
| integral to membrane |
| Component |
| cell part |
| membrane part |
| intrinsic to membrane |
| integral to membrane |
| Function |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides |
| Molecular Function |
| dihydroceramidase activity |
| Process |
| sphingoid metabolic process |
| metabolic process |
| ceramide metabolic process |
| sphingolipid metabolic process |
| membrane lipid metabolic process |
| primary metabolic process |
| lipid metabolic process |
| cellular lipid metabolic process |
|
| Cellular Location
|
- Endoplasmic reticulum membrane
- Multi-pass membrane protein
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Sphingolipid Metabolism | 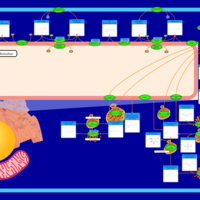 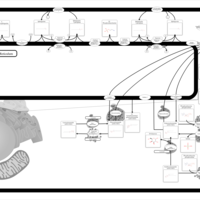 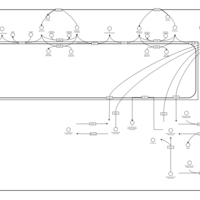 |  | | Gaucher Disease | 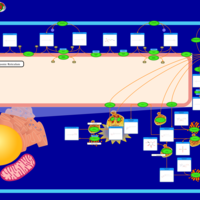   | Not Available | | Globoid Cell Leukodystrophy |   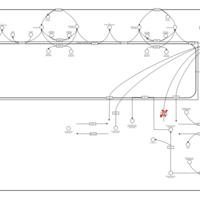 | Not Available | | Metachromatic Leukodystrophy (MLD) | 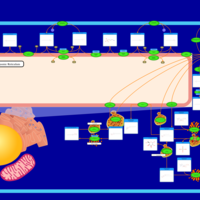 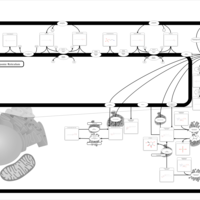 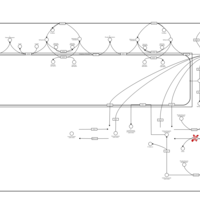 | Not Available | | Fabry disease |    | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 19 |
| Locus
| 19p13.3 |
| SNPs
| ACER1 |
| Gene Sequence
|
>795 bp
ATGCCTAGCATCTTCGCCTATCAGAGCTCCGAGGTGGACTGGTGTGAGAGCAACTTCCAG
TACTCGGAGCTGGTGGCCGAGTTCTACAACACGTTCTCCAATATCCCCTTCTTCATCTTC
GGGCCACTGATGATGCTCCTGATGCACCCGTATGCCCAGAAGCGCTCCCGCTACATTTAC
GTTGTCTGGGTCCTCTTCATGATCATAGGCCTGTTCTCCATGTATTTCCACATGACGCTC
AGCTTCCTGGGCCAGCTGCTGGACGAGATCGCCATCCTGTGGCTCCTGGGCAGTGGCTAT
AGCATATGGATGCCCCGCTGCTATTTCCCCTCCTTCCTTGGGGGGAACAGGTCCCAGTTC
ATCCGCCTGGTCTTCATCACCACTGTGGTCAGCACCCTTCTGTCCTTCCTGCGGCCCACG
GTCAACGCCTACGCCCTCAACAGCATTGCCCTGCACATTCTCTACATCGTGTGCCAGGAG
TACAGGAAGACCAGCAATAAGGAGCTTCGGCACCTGATTGAGGTCTCCGTGGTTTTATGG
GCTGTTGCTCTGACCAGCTGGATCAGTGACCGTCTGCTTTGCAGCTTCTGGCAGAGGATT
CATTTCTTCTATCTGCACAGCATCTGGCATGTGCTCATCAGCATCACCTTCCCTTATGGC
ATGGTCACCATGGCCTTGGTGGATGCCAACTATGAGATGCCAGGTGAAACCCTCAAAGTC
CGCTACTGGCCTCGGGACAGTTGGCCCGTGGGGCTGCCCTACGTGGAAATCCGGGGTGAT
GACAAGGACTGCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 264 |
| Molecular Weight
| 31095.165 |
| Theoretical pI
| 7.133 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Alkaline ceramidase 1
MPSIFAYQSSEVDWCESNFQYSELVAEFYNTFSNIPFFIFGPLMMLLMHPYAQKRSRYIY
VVWVLFMIIGLFSMYFHMTLSFLGQLLDEIAILWLLGSGYSIWMPRCYFPSFLGGNRSQF
IRLVFITTVVSTLLSFLRPTVNAYALNSIALHILYIVCQEYRKTSNKELRHLIEVSVVLW
AVALTSWISDRLLCSFWQRIHFFYLHSIWHVLISITFPYGMVTMALVDANYEMPGETLKV
RYWPRDSWPVGLPYVEIRGDDKDC
|
| External Links |
|---|
| GenBank ID Protein
| 19070367 |
| UniProtKB/Swiss-Prot ID
| Q8TDN7 |
| UniProtKB/Swiss-Prot Entry Name
| ACER1_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| AF347024 |
| GeneCard ID
| ACER1 |
| GenAtlas ID
| ACER1 |
| HGNC ID
| HGNC:18356 |
| References |
|---|
| General References
| Not Available |
