| Identification |
|---|
| HMDB Protein ID
| CDBP04427 |
| Secondary Accession Numbers
| Not Available |
| Name
| Ubiquitin-conjugating enzyme E2 W |
| Description
| Not Available |
| Synonyms
|
- SubName: Ubiquitin-conjugating enzyme E2W (Putative), isoform CRA_c
- Ubiquitin carrier protein W
- Ubiquitin-conjugating enzyme 16
- Ubiquitin-protein ligase W
- UBC-16
|
| Gene Name
| UBE2W |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acid-amino acid ligase activity |
| Specific Function
| Accepts ubiquitin from the E1 complex and catalyzes its covalent attachment to other proteins. Catalyzes monoubiquitination. Involved in degradation of misfolded chaperone substrates by mediating monoubiquitination of STUB1/CHIP, leading to recruitment of ATXN3 to monoubiquitinated STUB1/CHIP, and restriction of the length of ubiquitin chain attached to STUB1/CHIP substrates by ATXN3. After UV irradiation, but not after mitomycin-C (MMC) treatment, acts as a specific E2 ubiquitin-conjugating enzyme for the Fanconi anemia complex by associating with E3 ubiquitin-protein ligase FANCL and catalyzing monoubiquitination of FANCD2, a key step in the DNA damage pathway. In vitro catalyzes 'Lys-11'-linked polyubiquitination.
|
| GO Classification
|
| Biological Process |
| DNA repair |
| protein K11-linked ubiquitination |
| protein monoubiquitination |
| cellular response to misfolded protein |
| misfolded or incompletely synthesized protein catabolic process |
| proteasomal ubiquitin-dependent protein catabolic process |
| Cellular Component |
| nucleus |
| Function |
| small conjugating protein ligase activity |
| ligase activity |
| ligase activity, forming carbon-nitrogen bonds |
| acid-amino acid ligase activity |
| catalytic activity |
| Molecular Function |
| ubiquitin-protein ligase activity |
| ATP binding |
| Process |
| metabolic process |
| regulation of protein metabolic process |
| macromolecule metabolic process |
| biological regulation |
| regulation of biological process |
| regulation of metabolic process |
| regulation of macromolecule metabolic process |
| post-translational protein modification |
| macromolecule modification |
| protein modification process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | protein ubiquitination | Not Available | Not Available | | Ubiquitin mediated proteolysis | Not Available | 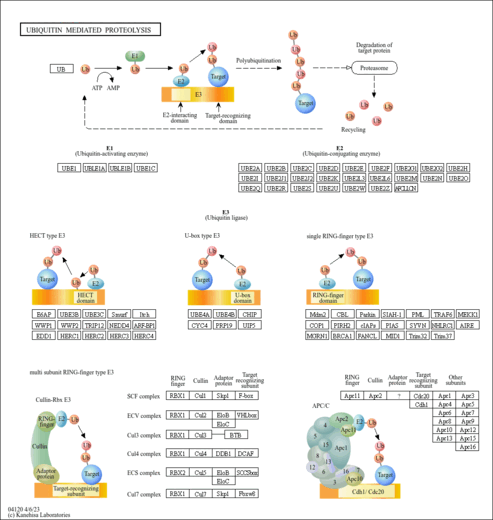 |
|
| Gene Properties |
|---|
| Chromosome Location
| 8 |
| Locus
| 8q21.11 |
| SNPs
| UBE2W |
| Gene Sequence
|
>489 bp
ATGGCGTCAATGCAGACCACAGGAAGGAGGGTGGAAGTATGGTTTCCAAAACGACTACAG
AAAGAACTGTTGGCTTTGCAAAATGACCCACCTCCTGGAATGACCTTAAATGAGAAGAGT
GTTCAAAATTCAATTACACAGTGGATTGTAGACATGGAAGGTGCACCAGGTACCTTATAT
GAAGGGGAAAAATTTCAACTTCTATTTAAATTTAGTAGTCGATATCCTTTTGACTCTCCT
CAGGTCATGTTTACTGGTGAAAATATTCCTGTTCATCCTCATGTTTATAGCAATGGTCAT
ATCTGTTTATCCATTCTAACAGAAGACTGGTCCCCAGCGCTCTCAGTCCAATCAGTTTGT
CTTAGCATTATTAGCATGCTTTCCAGCTGCAAGGAAAAGAGACGACCACCGGATAATTCT
TTTTATGTGCGAACATGTAACAAGAATCCAAAGAAAACAAAATGGTGGTATCATGATGAT
ACTTGTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 162 |
| Molecular Weight
| 22018.2 |
| Theoretical pI
| 9.148 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Ubiquitin-conjugating enzyme 16
MASMQTTGRRVEVWFPKRLQKELLALQNDPPPGMTLNEKSVQNSITQWIVDMEGAPGTLY
EGEKFQLLFKFSSRYPFDSPQVMFTGENIPVHPHVYSNGHICLSILTEDWSPALSVQSVC
LSIISMLSSCKEKRRPPDNSFYVRTCNKNPKKTKWWYHDDTC
|
| External Links |
|---|
| GenBank ID Protein
| 62997235 |
| UniProtKB/Swiss-Prot ID
| Q96B02 |
| UniProtKB/Swiss-Prot Entry Name
| Q1XBE0_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AY948289 |
| GeneCard ID
| UBE2W |
| GenAtlas ID
| UBE2W |
| HGNC ID
| HGNC:25616 |
| References |
|---|
| General References
| Not Available |
