| Identification |
|---|
| HMDB Protein ID
| CDBP04291 |
| Secondary Accession Numbers
| Not Available |
| Name
| Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- Dehydrogenase E1 and transketolase domain-containing protein 1
|
| Gene Name
| DHTKD1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in oxoglutarate dehydrogenase (succinyl-transferring) activity |
| Specific Function
| The 2-oxoglutarate dehydrogenase complex catalyzes the overall conversion of 2-oxoglutarate to succinyl-CoA and CO(2). It contains multiple copies of three enzymatic components: 2-oxoglutarate dehydrogenase (E1), dihydrolipoamide succinyltransferase (E2) and lipoamide dehydrogenase (E3) (By similarity).
|
| GO Classification
|
| Biological Process |
| glycolysis |
| tricarboxylic acid cycle |
| Cellular Component |
| mitochondrion |
| Function |
| binding |
| catalytic activity |
| vitamin binding |
| oxidoreductase activity |
| thiamin pyrophosphate binding |
| oxoglutarate dehydrogenase (succinyl-transferring) activity |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors |
| oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor |
| Molecular Function |
| oxoglutarate dehydrogenase (succinyl-transferring) activity |
| thiamine pyrophosphate binding |
| Process |
| metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| hexose metabolic process |
| glucose metabolic process |
| glucose catabolic process |
| glycolysis |
|
| Cellular Location
|
- Mitochondrion (Potential)
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Lysine Degradation | 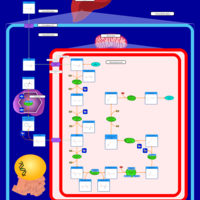 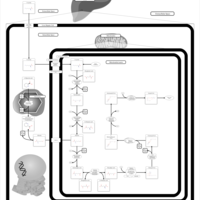 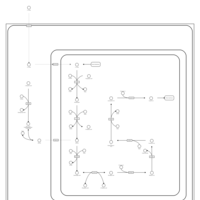 |  | | Saccharopinuria/Hyperlysinemia II | 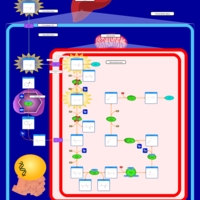 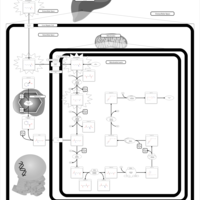 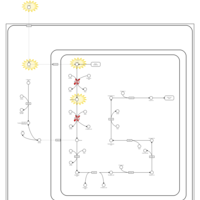 | Not Available | | Hyperlysinemia I, Familial | 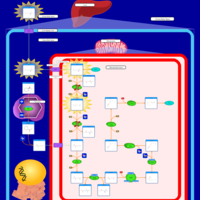 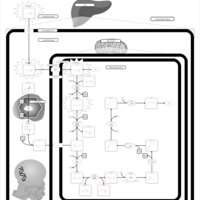 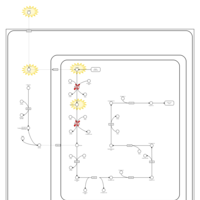 | Not Available | | Hyperlysinemia II or Saccharopinuria | 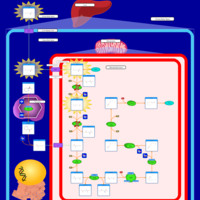 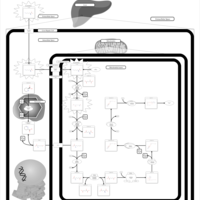  | Not Available | | Pyridoxine dependency with seizures |  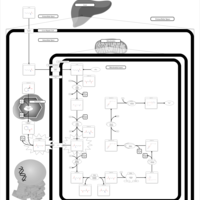  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 10 |
| Locus
| 10p14 |
| SNPs
| DHTKD1 |
| Gene Sequence
|
>2760 bp
ATGGCCTCTGCTACTGCGGCAGCAGCACGACGGGGCCTCGGCCGGGCTCTCCCTCTCCTC
TGGCGTGGCTACCAGACCGAGCGGGGCGTTTACGGCTACCGGCCGAGGAAGCCCGAGAGC
CGCGAGCCCCAGGGCGCCCTGGAGCGCCCCCCAGTTGATCATGGCCTTGCCAGGTTGGTG
ACAGTATATTGTGAGCATGGTCATAAAGCTGCCAAAATCAACCCCCTCTTCACCGGACAA
GCCCTGCTGGAGAATGTGCCTGAAATCCAAGCCCTGGTGCAGACACTGCAGGGACCCTTC
CACACGGCAGGATTATTGAACATGGGGAAGGAAGAGGCCTCACTTGAGGAAGTGTTAGTC
TATCTCAATCAAATCTACTGTGGGCAGATTTCTATTGAAACCTCCCAACTTCAGAGCCAG
GATGAGAAAGACTGGTTTGCCAAGCGGTTTGAGGAACTGCAAAAGGAGACGTTTACCACA
GAAGAGCGAAAACATCTGTCGAAACTAATGCTGGAATCTCAGGAGTTTGACCACTTTCTG
GCCACCAAGTTCTCGACAGTGAAGCGATATGGAGGCGAAGGGGCTGAAAGCATGATGGGC
TTTTTCCACGAGCTGCTGAAAATGTCGGCCTACAGCGGGATCACTGATGTCATTATTGGG
ATGCCCCATAGAGGGAGGCTGAATTTATTGACAGGCCTTCTGCAGTTCCCTCCAGAGCTG
ATGTTCCGTAAAATGCGAGGCTTAAGTGAATTTCCAGAGAATTTCTCAGCCACTGGAGAC
GTCCTGTCTCACCTGACCTCCTCTGTGGACCTGTACTTTGGGGCGCACCATCCCCTCCAT
GTGACAATGTTGCCCAATCCCTCGCACCTGGAGGCCGTCAACCCCGTGGCCGTGGGCAAA
ACTCGCGGCAGGCAGCAGTCTCGCCAAGACGGCGATTACTCTCCAGACAACTCAGCCCAG
CCGGGGGACAGGGTCATTTGCTTACAGGTCCATGGTGATGCTTCTTTCTGTGGTCAAGGG
ATTGTTCCTGAAACATTCACGCTGTCCAATCTCCCACATTTCAGAATTGGTGGGAGTGTG
CATTTGATTGTTAATAACCAGCTGGGTTACACCACTCCAGCTGAAAGAGGAAGGTCTTCT
TTATACTGCAGTGATATTGGGAAGCTTGTGGGCTGTGCCATCATCCATGTCAATGGAGAC
AGCCCAGAGGAAGTGGTCCGTGCCACACGACTGGCTTTTGAATACCAACGCCAGTTCCGC
AAGGATGTGATTATTGATCTGTTGTGCTACAGGCAGTGGGGCCACAATGAGCTGGATGAG
CCATTCTACACCAACCCCATCATGTACAAAATCATCAGAGCTCGAAAGAGCATTCCAGAC
ACATATGCAGAGCACCTCATTGCTGGCGGACTCATGACGCAGGAGGAGGTGTCTGAAATA
AAATCCTCCTACTATGCCAAGTTGAATGATCACTTAAATAACATGGCCCACTACAGGCCC
CCTGCCCTGAACCTGCAGGCCCACTGGCAGGGCCTGGCTCAGCCAGAAGCGCAAATCACC
ACCTGGAGTACAGGTGTGCCCCTCGACCTCCTGCGGTTTGTTGGCATGAAGTCTGTAGAG
GTGCCAAGAGAGCTGCAGATGCACAGTCACCTGCTGAAGACACATGTTCAGTCCAGAATG
GAGAAGATGATGGACGGAATCAAGCTAGACTGGGCCACCGCGGAAGCTCTTGCCTTGGGT
TCTTTACTTGCTCAAGGTTTTAATGTTCGTCTAAGTGGCCAAGATGTTGGTCGTGGAACT
TTCAGTCAGAGGCATGCAATCGTGGTTTGCCAGGAGACGGATGACACCTACATCCCCCTG
AACCATATGGACCCAAATCAGAAGGGGTTTCTAGAGGTCAGCAACAGCCCCCTGTCAGAA
GAGGCCGTCCTGGGATTCGAATATGGGATGAGCATTGAGAGCCCAAAGTTACTGCCCCTG
TGGGAGGCACAGTTTGGCGATTTCTTCAATGGTGCCCAGATCATCTTTGACACATTCATC
TCTGGAGGAGAGGCCAAGTGGCTCCTACAAAGCGGCATCGTCATCCTCCTTCCACATGGC
TACGATGGGGCTGGGCCAGACCACTCATCCTGTCGAATAGAGCGTTTCCTGCAGATGTGT
GACAGTGCGGAAGAGGGGGTGGACGGAGACACTGTGAACATGTTTGTGGTTCACCCAACA
ACTCCTGCACAGTATTTCCACTTGCTTAGGAGACAGATGGTCCGGAACTTCAGAAAACCA
CTCATTGTTGCTTCCCCTAAGATGTTACTCAGGCTCCCGGCAGCCGTGTCAACTCTTCAA
GAAATGGCACCAGGAACAACATTTAACCCGGTCATTGGTGATTCATCTGTGGATCCAAAA
AAGGTTAAGACCCTCGTGTTCTGCTCCGGCAAACATTTCTACTCCCTGGTGAAACAAAGA
GAATCTCTGGGGGCCAAGAAGCATGACTTTGCCATCATCCGAGTAGAGGAACTCTGCCCC
TTCCCGTTGGATTCTTTACAGCAAGAGATGAGCAAATACAAACATGTTAAAGATCATATT
TGGAGTCAGGAGGAACCTCAGAACATGGGTCCGTGGTCGTTTGTTTCTCCAAGGTTTGAA
AAGCAGCTGGCCTGCAAGCTCCGTCTGGTGGGCCGGCCCCCTTTGCCAGTACCCGCTGTA
GGAATTGGCACAGTTCACTTGCACCAGCATGAAGATATCCTCGCCAAGACCTTCGCTTGA
|
| Protein Properties |
|---|
| Number of Residues
| 919 |
| Molecular Weight
| 103042.15 |
| Theoretical pI
| 6.936 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial
MASATAAAARRGLGRALPLFWRGYQTERGVYGYRPRKPESREPQGALERPPVDHGLARLV
TVYCEHGHKAAKINPLFTGQALLENVPEIQALVQTLQGPFHTAGLLNMGKEEASLEEVLV
YLNQIYCGQISIETSQLQSQDEKDWFAKRFEELQKETFTTEERKHLSKLMLESQEFDHFL
ATKFSTVKRYGGEGAESMMGFFHELLKMSAYSGITDVIIGMPHRGRLNLLTGLLQFPPEL
MFRKMRGLSEFPENFSATGDVLSHLTSSVDLYFGAHHPLHVTMLPNPSHLEAVNPVAVGK
TRGRQQSRQDGDYSPDNSAQPGDRVICLQVHGDASFCGQGIVPETFTLSNLPHFRIGGSV
HLIVNNQLGYTTPAERGRSSLYCSDIGKLVGCAIIHVNGDSPEEVVRATRLAFEYQRQFR
KDVIIDLLCYRQWGHNELDEPFYTNPIMYKIIRARKSIPDTYAEHLIAGGLMTQEEVSEI
KSSYYAKLNDHLNNMAHYRPPALNLQAHWQGLAQPEAQITTWSTGVPLDLLRFVGMKSVE
VPRELQMHSHLLKTHVQSRMEKMMDGIKLDWATAEALALGSLLAQGFNVRLSGQDVGRGT
FSQRHAIVVCQETDDTYIPLNHMDPNQKGFLEVSNSPLSEEAVLGFEYGMSIESPKLLPL
WEAQFGDFFNGAQIIFDTFISGGEAKWLLQSGIVILLPHGYDGAGPDHSSCRIERFLQMC
DSAEEGVDGDTVNMFVVHPTTPAQYFHLLRRQMVRNFRKPLIVASPKMLLRLPAAVSTLQ
EMAPGTTFNPVIGDSSVDPKKVKTLVFCSGKHFYSLVKQRESLGAKKHDFAIIRVEELCP
FPLDSLQQEMSKYKHVKDHIWSQEEPQNMGPWSFVSPRFEKQLACKLRLVGRPPLPVPAV
GIGTVHLHQHEDILAKTFA
|
| External Links |
|---|
| GenBank ID Protein
| 38788380 |
| UniProtKB/Swiss-Prot ID
| Q96HY7 |
| UniProtKB/Swiss-Prot Entry Name
| DHTK1_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| NM_018706.5 |
| GeneCard ID
| DHTKD1 |
| GenAtlas ID
| DHTKD1 |
| HGNC ID
| HGNC:23537 |
| References |
|---|
| General References
| Not Available |
