| Identification |
|---|
| HMDB Protein ID
| CDBP03991 |
| Secondary Accession Numbers
| Not Available |
| Name
| Diphosphoinositol polyphosphate phosphohydrolase 1 |
| Description
| Not Available |
| Synonyms
|
- DIPP-1
- Diadenosine 5',5'''-P1,P6-hexaphosphate hydrolase 1
- Nucleoside diphosphate-linked moiety X motif 3
- Nudix motif 3
|
| Gene Name
| NUDT3 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity |
| Specific Function
| Cleaves a beta-phosphate from the diphosphate groups in PP-InsP5 (diphosphoinositol pentakisphosphate) and [PP]2-InsP4 (bisdiphosphoinositol tetrakisphosphate), suggesting that it may play a role in signal transduction. InsP6 (inositol hexakisphophate) is not a substrate. Acts as a negative regulator of the ERK1/2 pathway. Also able to catalyze the hydrolysis of dinucleoside oligophosphates, with Ap6A and Ap5A being the preferred substrates. The major reaction products are ADP and p4a from Ap6A and ADP and ATP from Ap5A. Also able to hydrolyze 5-phosphoribose 1-diphosphate.
|
| GO Classification
|
| Biological Process |
| cell-cell signaling |
| diadenosine polyphosphate catabolic process |
| diphosphoinositol polyphosphate catabolic process |
| Cellular Component |
| cytoplasm |
| Function |
| catalytic activity |
| hydrolase activity |
| Molecular Function |
| inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity |
| inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity |
| inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity |
| magnesium ion binding |
| diphosphoinositol-polyphosphate diphosphatase activity |
| inositol diphosphate tetrakisphosphate diphosphatase activity |
| inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity |
| inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity |
| inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity |
|
| Cellular Location
|
- Cytoplasm (Probable)
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Inositol Phosphate Metabolism | 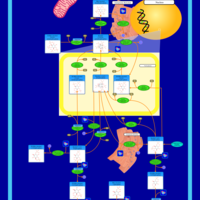 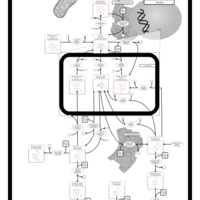 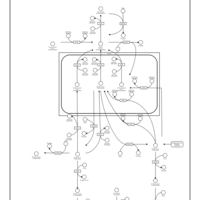 |  |
|
| Gene Properties |
|---|
| Chromosome Location
| 6 |
| Locus
| 6p21.2 |
| SNPs
| NUDT3 |
| Gene Sequence
|
>519 bp
ATGATGAAGCTCAAGTCGAACCAGACCCGCACCTACGACGGCGACGGCTACAAGAAGCGG
GCCGCATGCCTGTGTTTCCGCAGCGAGAGCGAGGAGGAGGTGCTACTCGTGAGCAGTAGT
CGCCATCCAGACAGATGGATTGTCCCTGGAGGAGGCATGGAGCCCGAGGAGGAGCCAAGT
GTGGCAGCAGTTCGTGAAGTCTGTGAGGAGGCTGGAGTAAAAGGGACATTGGGAAGATTA
GTTGGAATTTTTGAGAACCAGGAGAGGAAGCACAGGACGTATGTCTATGTGCTCATTGTC
ACTGAAGTGCTGGAAGACTGGGAAGATTCAGTTAACATTGGAAGGAAGAGGGAATGGTTT
AAAATAGAAGACGCCATAAAAGTGCTGCAGTATCACAAACCCGTGCAGGCATCATATTTT
GAAACATTGAGGCAAGGCTACTCAGCCAACAATGGCACCCCAGTCGTGGCCACCACATAC
TCGGTTTCTGCTCAGAGCTCGATGTCAGGCATCAGATGA
|
| Protein Properties |
|---|
| Number of Residues
| 172 |
| Molecular Weight
| 19470.855 |
| Theoretical pI
| 6.345 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Diphosphoinositol polyphosphate phosphohydrolase 1
MMKLKSNQTRTYDGDGYKKRAACLCFRSESEEEVLLVSSSRHPDRWIVPGGGMEPEEEPS
VAAVREVCEEAGVKGTLGRLVGIFENQERKHRTYVYVLIVTEVLEDWEDSVNIGRKREWF
KIEDAIKVLQYHKPVQASYFETLRQGYSANNGTPVVATTYSVSAQSSMSGIR
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| O95989 |
| UniProtKB/Swiss-Prot Entry Name
| NUDT3_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF062529 |
| GeneCard ID
| NUDT3 |
| GenAtlas ID
| NUDT3 |
| HGNC ID
| HGNC:8050 |
| References |
|---|
| General References
| Not Available |
