| Identification |
|---|
| HMDB Protein ID
| CDBP02605 |
| Secondary Accession Numbers
| Not Available |
| Name
| RuvB-like 2 |
| Description
| Not Available |
| Synonyms
|
- 48 kDa TATA box-binding protein-interacting protein
- 48 kDa TBP-interacting protein
- 51 kDa erythrocyte cytosolic protein
- ECP-51
- INO80 complex subunit J
- Repressing pontin 52
- Reptin 52
- TAP54-beta
- TIP49b
- TIP60-associated protein 54-beta
|
| Gene Name
| RUVBL2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in nucleotide binding |
| Specific Function
| Possesses single-stranded DNA-stimulated ATPase and ATP-dependent DNA helicase (5' to 3') activity; hexamerization is thought to be critical for ATP hydrolysis and adjacent subunits in the ring-like structure contribute to the ATPase activity.
Component of the NuA4 histone acetyltransferase complex which is involved in transcriptional activation of select genes principally by acetylation of nucleosomal histones H4 and H2A. This modification may both alter nucleosome - DNA interactions and promote interaction of the modified histones with other proteins which positively regulate transcription. This complex may be required for the activation of transcriptional programs associated with oncogene and proto-oncogene mediated growth induction, tumor suppressor mediated growth arrest and replicative senescence, apoptosis, and DNA repair. The NuA4 complex ATPase and helicase activities seem to be, at least in part, contributed by the association of RUVBL1 and RUVBL2 with EP400. NuA4 may also play a direct role in DNA repair when recruited to sites of DNA damage.
Proposed core component of the chromatin remodeling INO80 complex which is involved in transcriptional regulation, DNA replication and probably DNA repair.
Plays an essential role in oncogenic transformation by MYC and also modulates transcriptional activation by the LEF1/TCF1-CTNNB1 complex. May also inhibit the transcriptional activity of ATF2.
|
| GO Classification
|
| Biological Process |
| regulation of transcription, DNA-dependent |
| transcription, DNA-dependent |
| regulation of growth |
| DNA recombination |
| histone H2A acetylation |
| protein folding |
| DNA repair |
| histone H4 acetylation |
| cellular response to UV |
| Cellular Component |
| NuA4 histone acetyltransferase complex |
| cytoplasm |
| nuclear matrix |
| Ino80 complex |
| MLL1 complex |
| membrane |
| ribonucleoprotein complex |
| Function |
| purine nucleoside binding |
| adenyl nucleotide binding |
| adenyl ribonucleotide binding |
| atp binding |
| dna helicase activity |
| damaged dna binding |
| nucleic acid binding |
| dna binding |
| nucleoside-triphosphatase activity |
| helicase activity |
| hydrolase activity, acting on acid anhydrides |
| binding |
| hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| nucleotide binding |
| pyrophosphatase activity |
| catalytic activity |
| hydrolase activity |
| nucleoside binding |
| Molecular Function |
| damaged DNA binding |
| identical protein binding |
| unfolded protein binding |
| ATP binding |
| ATP-dependent DNA helicase activity |
| Process |
| metabolic process |
| macromolecule metabolic process |
| cellular macromolecule metabolic process |
| dna metabolic process |
| dna repair |
|
| Cellular Location
|
- Nucleus
- Cytoplasm
- Membrane
- nucleoplasm
- Nucleus matrix
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Pterine Biosynthesis |    | 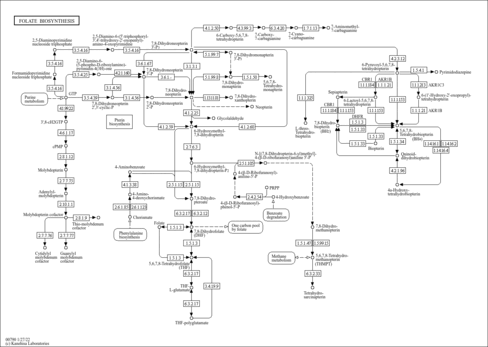 |
|
| Gene Properties |
|---|
| Chromosome Location
| 19 |
| Locus
| 19q13.3 |
| SNPs
| RUVBL2 |
| Gene Sequence
|
>1392 bp
ATGGCAACCGTTACAGCCACAACCAAAGTCCCGGAGATCCGTGATGTAACAAGGATTGAG
CGAATCGGTGCCCACTCCCACATCCGGGGACTGGGGCTGGACGATGCCTTGGAGCCTCGG
CAGGCTTCGCAAGGCATGGTGGGTCAGCTGGCGGCACGGCGGGCGGCTGGCGTGGTGCTG
GAGATGATCCGGGAAGGGAAGATTGCCGGTCGGGCAGTCCTTATTGCTGGCCAGCCGGGC
ACGGGGAAGACGGCCATCGCCATGGGCATGGCGCAGGCCCTGGGCCCTGACACGCCATTC
ACAGCCATCGCCGGCAGTGAAATCTTCTCCCTGGAGATGAGCAAGACCGAGGCGCTGACG
CAGGCCTTCCGGCGGTCCATCGGCGTTCGCATCAAGGAGGAGACGGAGATCATCGAAGGG
GAGGTGGTGGAGATCCAGATTGATCGACCAGCAACAGGGACGGGCTCCAAGGTGGGCAAA
CTGACCCTCAAGACCACAGAGATGGAGACCATCTACGACCTGGGCACCAAGATGATTGAG
TCCCTGACCAAGGACAAGGTCCAGGCCGGGGACGTGATCACCATCGACAAGGCGACGGGC
AAGATCTCCAAGCTGGGCCGCTCCTTCACACGCGCCCGCGACTACGACGCTATGGGCTCC
CAGACCAAGTTCGTGCAGTGCCCAGATGGGGAGCTCCAGAAACGCAAGGAGGTGGTGCAC
ACCGTGTCCCTGCACGAGATCGACGTCATCAACTCTCGCACCCAGGGCTTCCTGGCGCTC
TTCTCAGGTGACACAGGGGAGATCAAGTCAGAAGTCCGTGAGCAGATCAATGCCAAGGTG
GCTGAGTGGCGCGAGGAGGGCAAGGCGGAGATCATCCCTGGAGTGCTGTTCATCGACGAG
GTCCACATGCTGGACATCGAGAGCTTCTCCTTCCTCAACCGGGCCCTGGAGAGTGACATG
GCGCCTGTCCTGATCATGGCCACCAACCGTGGCATCACGCGAATCCGGGGCACCAGCTAC
CAGAGCCCTCACGGCATCCCCATAGACCTGCTGGACCGGCTGCTTATCGTCTCCACCACC
CCCTACAGCGAGAAAGACACGAAGCAGATCCTCCGCATCCGGTGCGAGGAAGAAGATGTG
GAGATGAGTGAGGACGCCTACACGGTGCTGACCCGCATCGGGCTGGAGACGTCACTGCGC
TACGCCATCCAGCTCATCACAGCTGCCAGCTTGGTGTGCCGGAAACGCAAGGGTACAGAA
GTGCAGGTGGATGACATCAAGCGGGTCTACTCACTCTTCCTGGACGAGTCCCGCTCCACG
CAGTACATGAAGGAGTACCAGGACGCCTTCCTCTTCAACGAACTCAAAGGCGAGACCATG
GACACCTCCTGA
|
| Protein Properties |
|---|
| Number of Residues
| 463 |
| Molecular Weight
| 51156.08 |
| Theoretical pI
| 5.642 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>RuvB-like 2
MATVTATTKVPEIRDVTRIERIGAHSHIRGLGLDDALEPRQASQGMVGQLAARRAAGVVL
EMIREGKIAGRAVLIAGQPGTGKTAIAMGMAQALGPDTPFTAIAGSEIFSLEMSKTEALT
QAFRRSIGVRIKEETEIIEGEVVEIQIDRPATGTGSKVGKLTLKTTEMETIYDLGTKMIE
SLTKDKVQAGDVITIDKATGKISKLGRSFTRARDYDAMGSQTKFVQCPDGELQKRKEVVH
TVSLHEIDVINSRTQGFLALFSGDTGEIKSEVREQINAKVAEWREEGKAEIIPGVLFIDE
VHMLDIESFSFLNRALESDMAPVLIMATNRGITRIRGTSYQSPHGIPIDLLDRLLIVSTT
PYSEKDTKQILRIRCEEEDVEMSEDAYTVLTRIGLETSLRYAIQLITAASLVCRKRKGTE
VQVDDIKRVYSLFLDESRSTQYMKEYQDAFLFNELKGETMDTS
|
| External Links |
|---|
| GenBank ID Protein
| 4587311 |
| UniProtKB/Swiss-Prot ID
| Q9Y230 |
| UniProtKB/Swiss-Prot Entry Name
| RUVB2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AB024301 |
| GeneCard ID
| RUVBL2 |
| GenAtlas ID
| RUVBL2 |
| HGNC ID
| HGNC:10475 |
| References |
|---|
| General References
| Not Available |
