| Identification |
|---|
| HMDB Protein ID
| CDBP02477 |
| Secondary Accession Numbers
| Not Available |
| Name
| Uridine phosphorylase 2 |
| Description
| Not Available |
| Synonyms
|
- UPase 2
- UrdPase 2
|
| Gene Name
| UPP2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in transferase activity, transferring pentosyl groups |
| Specific Function
| Catalyzes the reversible phosphorylytic cleavage of uridine and deoxyuridine to uracil and ribose- or deoxyribose-1-phosphate. The produced molecules are then utilized as carbon and energy sources or in the rescue of pyrimidine bases for nucleotide synthesis. Shows substrate specificity and accept uridine, deoxyuridine, and thymidine as well as the two pyrimidine nucleoside analogs 5-fluorouridine and 5-fluoro-2(')-deoxyuridine as substrates.
|
| GO Classification
|
| Biological Process |
| pyrimidine nucleoside salvage |
| pyrimidine nucleobase metabolic process |
| pyrimidine nucleoside catabolic process |
| UMP salvage |
| nucleotide catabolic process |
| uridine metabolic process |
| Cellular Component |
| cytosol |
| type III intermediate filament |
| Component |
| cell part |
| intracellular part |
| cytoplasm |
| Function |
| uridine phosphorylase activity |
| catalytic activity |
| transferase activity |
| transferase activity, transferring pentosyl groups |
| transferase activity, transferring glycosyl groups |
| Molecular Function |
| uridine phosphorylase activity |
| Process |
| nucleotide catabolic process |
| nucleoside metabolic process |
| metabolic process |
| nitrogen compound metabolic process |
| cellular nitrogen compound metabolic process |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolic process |
| nucleobase, nucleoside and nucleotide metabolic process |
| nucleoside phosphate metabolic process |
| nucleotide metabolic process |
|
| Cellular Location
|
Not Available
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | UMP biosynthesis via salvage pathway | Not Available | Not Available | | Drug metabolism - other enzymes | Not Available | 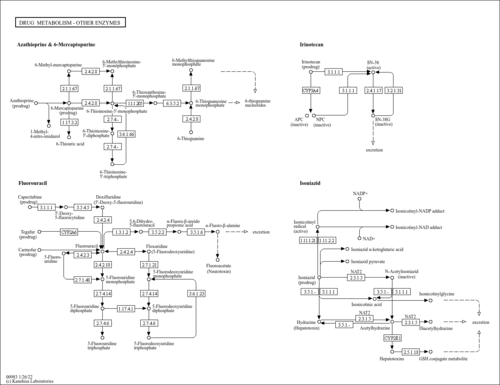 | | Pyrimidine Metabolism |    |  | | Beta Ureidopropionase Deficiency |    | Not Available | | UMP Synthase Deficiency (Orotic Aciduria) |  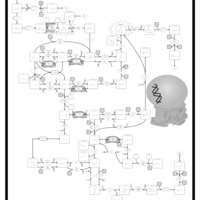 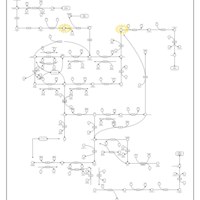 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 2 |
| Locus
| 2q24.1 |
| SNPs
| UPP2 |
| Gene Sequence
|
>954 bp
ATGGCTTCAGTTATACCTGCCTCCAATAGGTCCATGAGATCTGACAGGAATACATATGTT
GGAAAAAGGTTTGTTCACGTTAAAAATCCTTACTTGGATTTGATGGATGAAGACATTCTC
TATCACTTGGATTTGGGAACAAAAACACACAACCTACCAGCAATGTTTGGAGATGTAAAG
TTTGTCTGTGTCGGTGGGAGCCCCAACAGAATGAAAGCATTTGCACTGTTTATGCACAAG
GAGCTCGGGTTTGAGGAAGCTGAAGAAGACATAAAAGACATCTGTGCTGGGACAGACAGA
TACTGTATGTACAAAACCGGGCCTGTGCTCGCCATCAGTCACGGCATGGGCATCCCCTCC
ATTTCTATTATGCTTCATGAACTCATCAAATTACTCCACCATGCACGGTGCTGCGATGTC
ACCATTATTAGAATCGGTACATCAGGGGGAATAGGGATTGCACCAGGGACTGTTGTAATA
ACGGATATAGCTGTAGACTCCTTCTTTAAGCCCCGGTTTGAACAGGTCATTTTGGACAAC
ATTGTCACCCGAAGTACTGAACTGGACAAAGAACTGTCTGAAGAACTGTTCAACTGTAGC
AAAGAAATCCCCAACTTCCCAACCCTCGTTGGACATACAATGTGTACCTATGATTTTTAT
GAAGGCCAAGGCCGACTAGATGGAGCACTGTGCTCCTTTTCCAGAGAAAAAAAGTTAGAC
TACTTGAAGAGAGCATTTAAAGCTGGTGTCAGGAATATTGAAATGGAATCTACAGTGTTT
GCAGCTATGTGTGGACTCTGTGGTCTAAAAGCTGCTGTGGTCTGTGTGACACTTCTCGAC
AGACTCGACTGTGATCAGATCAACTTGCCTCATGATGTCCTGGTGGAGTACCAGCAACGG
CCTCAGCTCCTAATCTCCAACTTCATCAGACGGCGGCTTGGACTTTGTGACTAG
|
| Protein Properties |
|---|
| Number of Residues
| 317 |
| Molecular Weight
| 41601.795 |
| Theoretical pI
| 6.4 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Uridine phosphorylase 2
MASVIPASNRSMRSDRNTYVGKRFVHVKNPYLDLMDEDILYHLDLGTKTHNLPAMFGDVK
FVCVGGSPNRMKAFALFMHKELGFEEAEEDIKDICAGTDRYCMYKTGPVLAISHGMGIPS
ISIMLHELIKLLHHARCCDVTIIRIGTSGGIGIAPGTVVITDIAVDSFFKPRFEQVILDN
IVTRSTELDKELSEELFNCSKEIPNFPTLVGHTMCTYDFYEGQGRLDGALCSFSREKKLD
YLKRAFKAGVRNIEMESTVFAAMCGLCGLKAAVVCVTLLDRLDCDQINLPHDVLVEYQQR
PQLLISNFIRRRLGLCD
|
| External Links |
|---|
| GenBank ID Protein
| 4204340 |
| UniProtKB/Swiss-Prot ID
| O95045 |
| UniProtKB/Swiss-Prot Entry Name
| UPP2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AC005539 |
| GeneCard ID
| UPP2 |
| GenAtlas ID
| UPP2 |
| HGNC ID
| HGNC:23061 |
| References |
|---|
| General References
| Not Available |

