| Identification |
|---|
| HMDB Protein ID
| CDBP02439 |
| Secondary Accession Numbers
| Not Available |
| Name
| Isobutyryl-CoA dehydrogenase, mitochondrial |
| Description
| Not Available |
| Synonyms
|
- ACAD-8
- ARC42
- Activator-recruited cofactor 42 kDa component
- Acyl-CoA dehydrogenase family member 8
|
| Gene Name
| ACAD8 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in acyl-CoA dehydrogenase activity |
| Specific Function
| Has very high activity toward isobutyryl-CoA. Is an isobutyryl-CoA dehydrogenase that functions in valine catabolism. Plays a role in transcriptional coactivation within the ARC complex.
|
| GO Classification
|
| Biological Process |
| branched-chain amino acid catabolic process |
| cellular nitrogen compound metabolic process |
| lipid metabolic process |
| regulation of transcription, DNA-dependent |
| transcription, DNA-dependent |
| valine catabolic process |
| Cellular Component |
| mitochondrial matrix |
| Function |
| adenyl nucleotide binding |
| oxidoreductase activity |
| fad or fadh2 binding |
| oxidoreductase activity, acting on the ch-ch group of donors |
| acyl-coa dehydrogenase activity |
| binding |
| catalytic activity |
| nucleoside binding |
| purine nucleoside binding |
| Molecular Function |
| acyl-CoA dehydrogenase activity |
| flavin adenine dinucleotide binding |
| Process |
| metabolic process |
| oxidation reduction |
|
| Cellular Location
|
- Mitochondrion
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | L-valine degradation | Not Available | Not Available | | Valine, Leucine and Isoleucine Degradation |  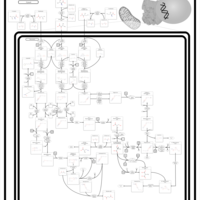 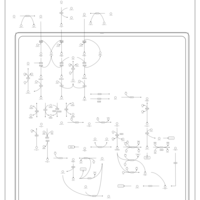 |  | | Beta-Ketothiolase Deficiency |  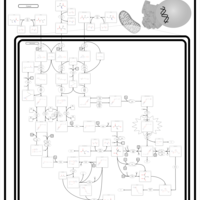 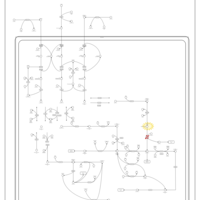 | Not Available | | 2-Methyl-3-Hydroxybutryl CoA Dehydrogenase Deficiency |  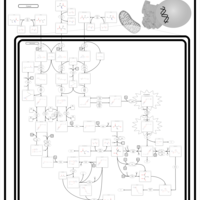 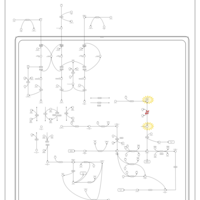 | Not Available | | Propionic Acidemia | 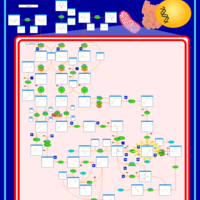 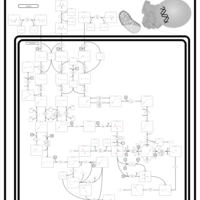 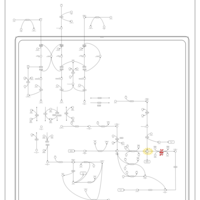 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 11 |
| Locus
| 11q25 |
| SNPs
| ACAD8 |
| Gene Sequence
|
>1248 bp
ATGCTGTGGAGCGGCTGCCGGCGTTTCGGGGCGCGCCTCGGCTGCCTGCCCGGCGGTCTC
CGGGTCCTCGTCCAGACCGGCCACCGGAGCTTGACCTCCTGCATCGACCCTTCCATGGGA
CTTAATGAAGAGCAGAAAGAATTTCAAAAAGTGGCCTTTGACTTTGCTGCCCGAGAGATG
GCTCCAAATATGGCAGAGTGGGACCAGAAGGAGCTGTTCCCAGTGGATGTGATGCGGAAG
GCAGCCCAGCTAGGCTTCGGAGGGGTCTACATACAAACAGATGTGGGCGGGTCTGGGCTG
TCACGTCTTGATACCTCTGTCATTTTTGAAGCCTTGGCTACAGGCTGCACCAGCACCACA
GCCTATATAAGCATCCACAACATGTGTGCCTGGATGATTGATAGCTTCGGAAATGAGGAA
CAGAGGCACAAATTTTGCCCACCGCTCTGTACCATGGAGAAGTTTGCTTCCTACTGCCTC
ACTGAACCAGGAAGTGGGAGTGATGCTGCCTCTCTTCTGACCTCCGCTAAGAAACAGGGA
GATCATTACATCCTCAATGGCTCCAAGGCCTTCATCAGTGGTGCTGGTGAGTCAGACATC
TATGTGGTCATGTGCCGAACAGGAGGACCAGGCCCCAAGGGCATCTCATGCATAGTTGTT
GAGAAGGGGACCCCTGGCCTCAGCTTTGGCAAGAAGGAGAAAAAGGTGGGGTGGAACTCC
CAGCCAACACGAGCTGTGATCTTCGAAGACTGTGCTGTCCCTGTGGCCAACAGAATTGGG
AGCGAGGGGCAGGGCTTCCTCATTGCCGTGAGAGGACTGAACGGAGGGAGGATCAATATT
GCTTCCTGCTCCCTGGGGGCTGCCCACGCCTCTGTCATCCTCACCCGAGACCACCTCAAT
GTCCGGAAGCAGTTTGGAGAGCCTCTGGCCAGTAACCAGTACTTGCAATTCACACTGGCT
GATATGGCAACAAGGCTGGTGGCCGCGCGGCTGATGGTCCGCAATGCAGCAGTGGCTCTG
CAGGAGGAGAGGAAGGATGCAGTGGCCTTGTGCTCCATGGCCAAGCTCTTTGCTACAGAT
GAATGCTTTGCCATCTGCAACCAGGCCTTGCAGATGCACGGGGGCTACGGCTACCTGAAG
GATTACGCTGTTCAGCAGTACGTGCGGGACTCCAGGGTCCACCAGATTCTAGAAGGTAGC
AATGAAGTGATGAGGATACTGATCTCTAGAAGCCTGCTTCAGGAGTAG
|
| Protein Properties |
|---|
| Number of Residues
| 415 |
| Molecular Weight
| 45069.39 |
| Theoretical pI
| 7.849 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Isobutyryl-CoA dehydrogenase, mitochondrial
MLWSGCRRFGARLGCLPGGLRVLVQTGHRSLTSCIDPSMGLNEEQKEFQKVAFDFAAREM
APNMAEWDQKELFPVDVMRKAAQLGFGGVYIQTDVGGSGLSRLDTSVIFEALATGCTSTT
AYISIHNMCAWMIDSFGNEEQRHKFCPPLCTMEKFASYCLTEPGSGSDAASLLTSAKKQG
DHYILNGSKAFISGAGESDIYVVMCRTGGPGPKGISCIVVEKGTPGLSFGKKEKKVGWNS
QPTRAVIFEDCAVPVANRIGSEGQGFLIAVRGLNGGRINIASCSLGAAHASVILTRDHLN
VRKQFGEPLASNQYLQFTLADMATRLVAARLMVRNAAVALQEERKDAVALCSMAKLFATD
ECFAICNQALQMHGGYGYLKDYAVQQYVRDSRVHQILEGSNEVMRILISRSLLQE
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q9UKU7 |
| UniProtKB/Swiss-Prot Entry Name
| ACAD8_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| AF126245 |
| GeneCard ID
| ACAD8 |
| GenAtlas ID
| ACAD8 |
| HGNC ID
| HGNC:87 |
| References |
|---|
| General References
| Not Available |
