| Identification |
|---|
| HMDB Protein ID
| CDBP02361 |
| Secondary Accession Numbers
| Not Available |
| Name
| Putative N-acetylglucosamine-6-phosphate deacetylase |
| Description
| Not Available |
| Synonyms
|
- Amidohydrolase domain-containing protein 2
- GlcNAc 6-P deacetylase
|
| Gene Name
| AMDHD2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in hydrolase activity |
| Specific Function
| Hydrolyzes the N-glycolyl group from N-glycolylglucosamine 6-phosphate (GlcNGc-6-P) in the N-glycolylneuraminic acid (Neu5Gc) degradation pathway. Although human is not able to catalyze formation of Neu5Gc due to the inactive CMAHP enzyme, Neu5Gc is present in food and must be degraded.
|
| GO Classification
|
| Biological Process |
| N-acetylneuraminate catabolic process |
| N-acetylglucosamine metabolic process |
| carbohydrate metabolic process |
| Function |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds |
| hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides |
| n-acetylglucosamine-6-phosphate deacetylase activity |
| Molecular Function |
| metal ion binding |
| N-acetylglucosamine-6-phosphate deacetylase activity |
| Process |
| metabolic process |
| small molecule metabolic process |
| alcohol metabolic process |
| monosaccharide metabolic process |
| amino sugar metabolic process |
| glucosamine metabolic process |
| n-acetylglucosamine metabolic process |
|
| Cellular Location
|
- Cytoplasmic
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Amino Sugar Metabolism | 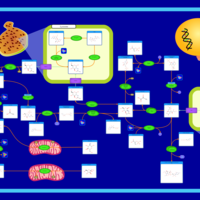 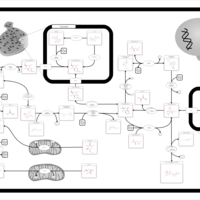  |  | | N-acetylneuraminate degradation | Not Available | Not Available | | Amino sugar and nucleotide sugar metabolism | Not Available |  | | Sialuria or French Type Sialuria | 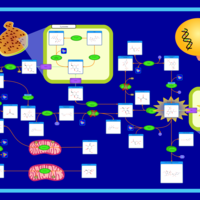 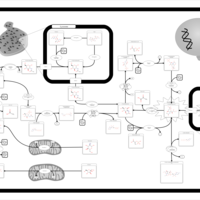 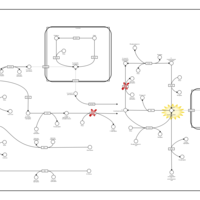 | Not Available | | Salla Disease/Infantile Sialic Acid Storage Disease | 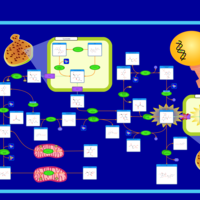 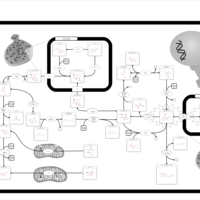  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 16 |
| Locus
| 16p13.3 |
| SNPs
| AMDHD2 |
| Gene Sequence
|
>1320 bp
ATGCGCGGCGAGCAGGGCGCGGCGGGGGCCCGCGTGCTCCAGTTCACTAACTGCCGGATC
CTGCGCGGAGGGAAACTGCTCAGGGAGGATCTGTGGGTGCGCGGAGGCCGCATCTTGGAC
CCAGAGAAGCTGTTCTTTGAGGAGCGGCGCGTGGCCGACGAGCGGCGGGACTGCGGGGGC
CGCATCTTGGCTCCCGGATTCATCGACGTGCAGATCAACGGTGGATTTGGTGTTGACTTC
TCTCAAGCCACGGAGGACGTGGGTTCGGGGGTTGCCCTCGTGGCCCGGAGGATCCTGTCG
CACGGCGTCACCTCCTTCTGCCCCACCCTGGTCACTTCCCCACCGGAGGTTTATCACAAG
GTTGTTCCTCAGATCCCTGTGAAGAGTGGTGGTCCCCATGGGGCAGGGGTCCTCGGGCTG
CACCTGGAGGGCCCCTTCATCAGCCGGGAGAAGCGGGGCGCGCACCCCGAGGCCCACCTC
CGCTCCTTCGAGGCCGATGCCTTCCAGGACTTGCTGGCCACCTACGGGCCCCTGGACAAT
GTCCGCATCGTGACGCTGGCCCCAGAGTTGGGCCGTAGCCACGAAGTGATCCGGGCGCTG
ACGGCCCGTGGCATCTGCGTGTCCCTAGGGCACTCAGTGGCTGACCTGCGGGCGGCAGAG
GATGCTGTGTGGAGCGGAGCCACCTTCATCACCCACCTCTTCAACGCCATGCTGCCTTTC
CACCACCGCGACCCAGGCATCGTGGGGCTCCTGACCAGCGACCGGCTGCCCGCAGGCCGC
TGCATCTTCTATGGGATGATTGCAGATGGCACGCACACCAACCCCGCCGCCCTGCGGATC
GCCCACCGTGCCCATCCCCAGGGGCTGGTGCTGGTCACCGATGCCATCCCTGCCTTGGGC
CTGGGCAACGGCCGGCACACGCTGGGACAGCAGGAAGTGGAAGTGGACGGTCTGACGGCC
TACGTGGCAGGTGAGCGCCCTGACCCACTGGGTCCCAGGTCCCAGCCCGCATGCCAGGTG
GCCCACGACCCCCCCAGAGCCTGCCCTCTCTGCTCTCAAGGCACCAAGACGCTGAGTGGC
AGCATAGCCCCAATGGACGTCTGTGTCCGGCACTTCCTGCAGGCCACAGGCTGCAGCATG
GAGTCGGCCCTGGAGGCTGCATCCCTGCACCCCGCCCAGTTGCTGGGGCTGGAGAAGAGT
AAGGGGACCCTGGACTTTGGTGCTGACGCAGACTTCGTGGTGCTCGACGACTCCCTTCAC
GTCCAGGCCACCTACATCTCGGGTGAGCTGGTGTGGCAGGCGGACGCAGCTAGGCAGTGA
|
| Protein Properties |
|---|
| Number of Residues
| 409 |
| Molecular Weight
| 63594.415 |
| Theoretical pI
| 7.526 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Putative N-acetylglucosamine-6-phosphate deacetylase
MRGEQGAAGARVLQFTNCRILRGGKLLREDLWVRGGRILDPEKLFFEERRVADERRDCGG
RILAPGFIDVQINGGFGVDFSQATEDVGSGVALVARRILSHGVTSFCPTLVTSPPEVYHK
VVPQIPVKSGGPHGAGVLGLHLEGPFISREKRGAHPEAHLRSFEADAFQDLLATYGPLDN
VRIVTLAPELGRSHEVIRALTARGICVSLGHSVADLRAAEDAVWSGATFITHLFNAMLPF
HHRDPGIVGLLTSDRLPAGRCIFYGMIADGTHTNPAALRIAHRAHPQGLVLVTDAIPALG
LGNGRHTLGQQEVEVDGLTAYVAGTKTLSGSIAPMDVCVRHFLQATGCSMESALEAASLH
PAQLLGLEKSKGTLDFGADADFVVLDDSLHVQATYISGELVWQADAARQ
|
| External Links |
|---|
| GenBank ID Protein
| 21361513 |
| UniProtKB/Swiss-Prot ID
| Q9Y303 |
| UniProtKB/Swiss-Prot Entry Name
| NAGA_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| Not Available |
| GeneCard ID
| AMDHD2 |
| GenAtlas ID
| AMDHD2 |
| HGNC ID
| HGNC:24262 |
| References |
|---|
| General References
| Not Available |
