| Identification |
|---|
| HMDB Protein ID
| CDBP01940 |
| Secondary Accession Numbers
| Not Available |
| Name
| E3 ubiquitin-protein ligase Mdm2 |
| Description
| Not Available |
| Synonyms
|
- Double minute 2 protein
- Hdm2
- Oncoprotein Mdm2
- p53-binding protein Mdm2
|
| Gene Name
| MDM2 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in negative regulation of apoptosis |
| Specific Function
| E3 ubiquitin-protein ligase that mediates ubiquitination of p53/TP53, leading to its degration by the proteasome. Inhibits p53/TP53- and p73/TP73-mediated cell cycle arrest and apoptosis by binding its transcriptional activation domain. Also acts as an ubiquitin ligase E3 toward itself and ARRB1. Permits the nuclear export of TP53/p53. Promotes proteasome-dependent ubiquitin- independent degradation of retinoblastoma RB1 protein. Inhibits DAXX-mediated apoptosis by inducing its ubiquitination and degradation |
| GO Classification
|
| Component |
| cell part |
| intracellular |
| Function |
| ion binding |
| cation binding |
| metal ion binding |
| binding |
| transition metal ion binding |
| zinc ion binding |
| protein binding |
|
| Cellular Location
|
- Nucleus
- Nucleus
- Cytoplasm
- nucleolus
- nucleoplasm
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | BCR-ABL Action in CML Pathogenesis | 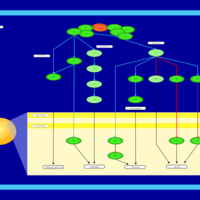 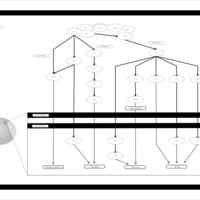 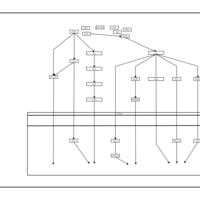 | Not Available | | Imatinib Inhibition of BCR-ABL | 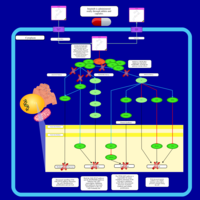 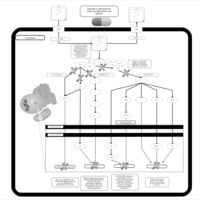  | Not Available | | Dasatinib Inhibition of BCR-ABL | 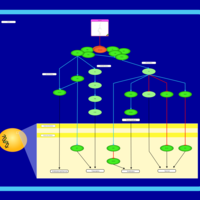 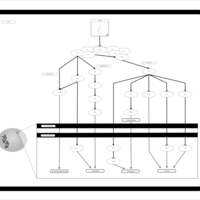 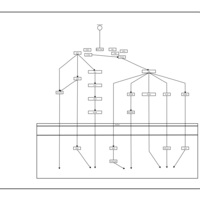 | Not Available | | Nilotinib Inhibition of BCR-ABL | 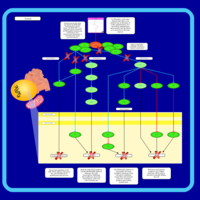 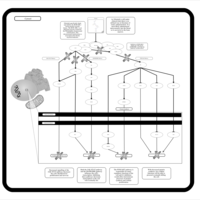 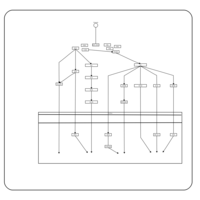 | Not Available | | Bosutinib Inhibition of BCR-ABL | 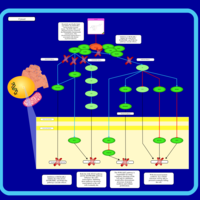 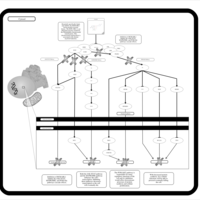 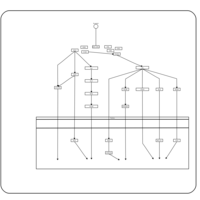 | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| Chromosome:1 |
| Locus
| 12q14.3-q15 |
| SNPs
| MDM2 |
| Gene Sequence
|
>1476 bp
ATGTGCAATACCAACATGTCTGTACCTACTGATGGTGCTGTAACCACCTCACAGATTCCA
GCTTCGGAACAAGAGACCCTGGTTAGACCAAAGCCATTGCTTTTGAAGTTATTAAAGTCT
GTTGGTGCACAAAAAGACACTTATACTATGAAAGAGGTTCTTTTTTATCTTGGCCAGTAT
ATTATGACTAAACGATTATATGATGAGAAGCAACAACATATTGTATATTGTTCAAATGAT
CTTCTAGGAGATTTGTTTGGCGTGCCAAGCTTCTCTGTGAAAGAGCACAGGAAAATATAT
ACCATGATCTACAGGAACTTGGTAGTAGTCAATCAGCAGGAATCATCGGACTCAGGTACA
TCTGTGAGTGAGAACAGGTGTCACCTTGAAGGTGGGAGTGATCAAAAGGACCTTGTACAA
GAGCTTCAGGAAGAGAAACCTTCATCTTCACATTTGGTTTCTAGACCATCTACCTCATCT
AGAAGGAGAGCAATTAGTGAGACAGAAGAAAATTCAGATGAATTATCTGGTGAACGACAA
AGAAAACGCCACAAATCTGATAGTATTTCCCTTTCCTTTGATGAAAGCCTGGCTCTGTGT
GTAATAAGGGAGATATGTTGTGAAAGAAGCAGTAGCAGTGAATCTACAGGGACGCCATCG
AATCCGGATCTTGATGCTGGTGTAAGTGAACATTCAGGTGATTGGTTGGATCAGGATTCA
GTTTCAGATCAGTTTAGTGTAGAATTTGAAGTTGAATCTCTCGACTCAGAAGATTATAGC
CTTAGTGAAGAAGGACAAGAACTCTCAGATGAAGATGATGAGGTATATCAAGTTACTGTG
TATCAGGCAGGGGAGAGTGATACAGATTCATTTGAAGAAGATCCTGAAATTTCCTTAGCT
GACTATTGGAAATGCACTTCATGCAATGAAATGAATCCCCCCCTTCCATCACATTGCAAC
AGATGTTGGGCCCTTCGTGAGAATTGGCTTCCTGAAGATAAAGGGAAAGATAAAGGGGAA
ATCTCTGAGAAAGCCAAACTGGAAAACTCAACACAAGCTGAAGAGGGCTTTGATGTTCCT
GATTGTAAAAAAACTATAGTGAATGATTCCAGAGAGTCATGTGTTGAGGAAAATGATGAT
AAAATTACACAAGCTTCACAATCACAAGAAAGTGAAGACTATTCTCAGCCATCAACTTCT
AGTAGCATTATTTATAGCAGCCAAGAAGATGTGAAAGAGTTTGAAAGGGAAGAAACCCAA
GACAAAGAAGAGAGTGTGGAATCTAGTTTGCCCCTTAATGCCATTGAACCTTGTGTGATT
TGTCAAGGTCGACCTAAAAATGGTTGCATTGTCCATGGCAAAACAGGACATCTTATGGCC
TGCTTTACATGTGCAAAGAAGCTAAAGAAAAGGAATAAGCCCTGCCCAGTATGTAGACAA
CCAATTCAAATGATTGTGCTAACTTATTTCCCCTAG
|
| Protein Properties |
|---|
| Number of Residues
| 491 |
| Molecular Weight
| 55232.4 |
| Theoretical pI
| 4.32 |
| Pfam Domain Function
|
|
| Signals
|
|
|
Transmembrane Regions
|
|
| Protein Sequence
|
>E3 ubiquitin-protein ligase Mdm2
MCNTNMSVPTDGAVTTSQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQY
IMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGT
SVSENRCHLEGGSDQKDLVQELQEEKPSSSHLVSRPSTSSRRRAISETEENSDELSGERQ
RKRHKSDSISLSFDESLALCVIREICCERSSSSESTGTPSNPDLDAGVSEHSGDWLDQDS
VSDQFSVEFEVESLDSEDYSLSEEGQELSDEDDEVYQVTVYQAGESDTDSFEEDPEISLA
DYWKCTSCNEMNPPLPSHCNRCWALRENWLPEDKGKDKGEISEKAKLENSTQAEEGFDVP
DCKKTIVNDSRESCVEENDDKITQASQSQESEDYSQPSTSSSIIYSSQEDVKEFEREETQ
DKEESVESSLPLNAIEPCVICQGRPKNGCIVHGKTGHLMACFTCAKKLKKRNKPCPVCRQ
PIQMIVLTYFP
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| Q00987 |
| UniProtKB/Swiss-Prot Entry Name
| MDM2_HUMAN |
| PDB IDs
|
|
| GenBank Gene ID
| M92424 |
| GeneCard ID
| MDM2 |
| GenAtlas ID
| MDM2 |
| HGNC ID
| HGNC:6973 |
| References |
|---|
| General References
| Not Available |