| Identification |
|---|
| HMDB Protein ID
| CDBP01610 |
| Secondary Accession Numbers
| Not Available |
| Name
| Epoxide hydrolase 1 |
| Description
| Not Available |
| Synonyms
|
- Epoxide hydratase
- Microsomal epoxide hydrolase
|
| Gene Name
| EPHX1 |
| Protein Type
| Enzyme |
| Biological Properties |
|---|
| General Function
| Involved in catalytic activity |
| Specific Function
| Biotransformation enzyme that catalyzes the hydrolysis of arene and aliphatic epoxides to less reactive and more water soluble dihydrodiols by the trans addition of water.
|
| GO Classification
|
| Biological Process |
| aromatic compound catabolic process |
| response to toxin |
| response to organic cyclic compound |
| Cellular Component |
| endoplasmic reticulum membrane |
| integral to membrane |
| Component |
| membrane |
| cell part |
| Function |
| catalytic activity |
| hydrolase activity |
| hydrolase activity, acting on ether bonds |
| ether hydrolase activity |
| epoxide hydrolase activity |
| Molecular Function |
| cis-stilbene-oxide hydrolase activity |
| epoxide hydrolase activity |
| Process |
| response to stimulus |
| response to chemical stimulus |
| response to toxin |
|
| Cellular Location
|
- Endoplasmic reticulum membrane
- Single-pass type II membrane protein
- Microsome membrane
- Single-pass type II membrane protein (Potential)
|
| Pathways
|
| Name | SMPDB/Pathwhiz | KEGG | | Metabolism of xenobiotics by cytochrome P450 | Not Available | 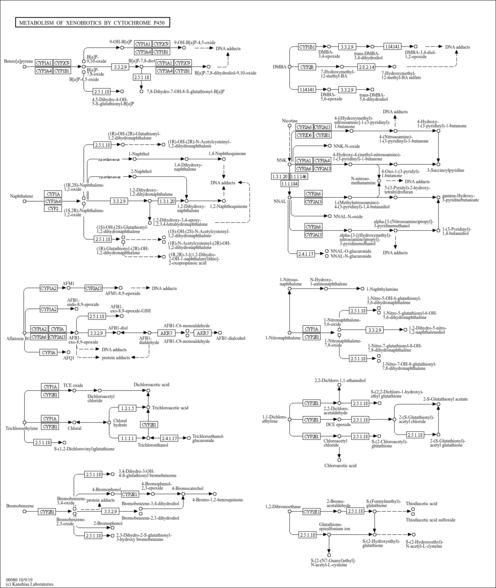 | | Bile secretion | Not Available | 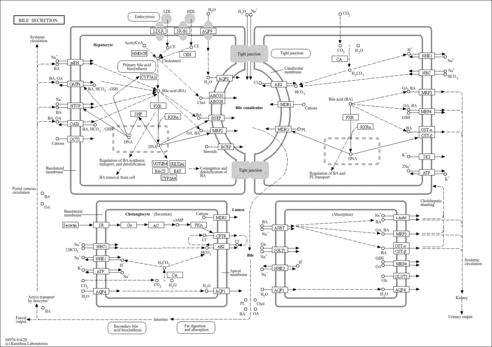 | | Chemical carcinogenesis | Not Available | 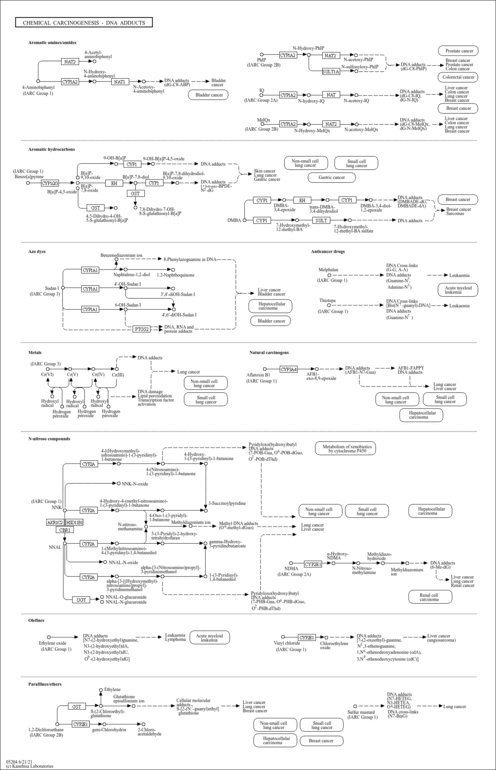 | | Phenytoin (Antiarrhythmic) Action Pathway |    | Not Available | | Carbamazepine Metabolism Pathway | 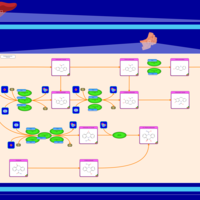 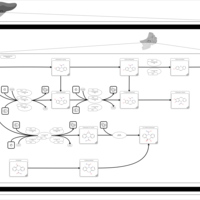  | Not Available |
|
| Gene Properties |
|---|
| Chromosome Location
| 1 |
| Locus
| 1q42.1 |
| SNPs
| EPHX1 |
| Gene Sequence
|
>1368 bp
ATGTGGCTAGAAATCCTCCTCACTTCAGTGCTGGGCTTTGCCATCTACTGGTTCATCTCC
CGGGACAAAGAGGAAACTTTGCCACTTGAAGATGGGTGGTGGGGGCCAGGCACGAGGTCC
GCAGCCAGGGAGGACGACAGCATCCGCCCTTTCAAGGTGGAAACGTCAGATGAGGAGATC
CACGACTTACACCAGAGGATCGATAAGTTCCGTTTCACCCCACCTTTGGAGGACAGCTGC
TTCCACTATGGCTTCAACTCCAACTACCTGAAGAAAGTCATCTCCTACTGGCGGAATGAA
TTTGACTGGAAGAAGCAGGTGGAGATTCTCAACAGATACCCTCACTTCAAGACTAAGATT
GAAGGGCTGGACATCCACTTCATCCACGTGAAGCCCCCCCAGCTGCCCGCAGGCCATACC
CCGAAGCCCTTGCTGATGGTGCACGGCTGGCCCGGCTCTTTCTACGAGTTTTATAAGATC
ATCCCACTCCTGACTGACCCCAAGAACCATGGCCTGAGCGATGAGCACGTTTTTGAAGTC
ATCTGCCCTTCCATCCCTGGCTATGGCTTCTCAGAGGCATCCTCCAAGAAGGGGTTCAAC
TCGGTGGCCACCGCCAGGATCTTTTACAAGCTGATGCTGCGGCTGGGCTTCCAGGAATTC
TACATTCAAGGAGGGGACTGGGGGTCCCTGATCTGCACTAATATGGCCCAGCTGGTGCCC
AGCCACGTGAAAGGCCTGCACTTGAACATGGCTTTGGTTTTAAGCAACTTCTCTACCCTG
ACCCTCCTCCTGGGACAGCGTTTCGGGAGGTTTCTTGGCCTCACTGAGAGGGATGTGGAG
CTGCTGTACCCCGTCAAGGAGAAGGTATTCTACAGCCTGATGAGGGAGAGCGGCTACATG
CACATCCAGTGCACCAAGCCTGACACCGTAGGCTCTGCTCTGAATGACTCTCCTGTGGGT
CTGGCTGCCTATATTCTAGAGAAGTTTTCCACCTGGACCAATACGGAATTCCGATACCTG
GAGGATGGAGGCCTGGAAAGGAAGTTCTCCCTGGACGACCTGCTGACCAACGTCATGCTC
TACTGGACAACAGGCACCATCATCTCCTCCCAGCGCTTCTACAAGGAGAACCTGGGACAG
GGCTGGATGACCCAGAAGCATGAGCGGATGAAGGTCTATGTGCCCACTGGCTTCTCTGCC
TTCCCTTTTGAGCTATTGCACACGCCTGAAAAGTGGGTGAGGTTCAAGTACCCAAAGCTC
ATCTCCTATTCCTACATGGTTCGTGGGGGCCACTTTGCGGCCTTTGAGGAGCCGGAGCTG
CTCGCCCAGGACATCCGCAAGTTCCTGTCGGTGCTGGAGCGGCAATGA
|
| Protein Properties |
|---|
| Number of Residues
| 455 |
| Molecular Weight
| 52948.48 |
| Theoretical pI
| 7.256 |
| Pfam Domain Function
|
|
| Signals
|
Not Available
|
|
Transmembrane Regions
|
Not Available
|
| Protein Sequence
|
>Epoxide hydrolase 1
MWLEILLTSVLGFAIYWFISRDKEETLPLEDGWWGPGTRSAAREDDSIRPFKVETSDEEI
HDLHQRIDKFRFTPPLEDSCFHYGFNSNYLKKVISYWRNEFDWKKQVEILNRYPHFKTKI
EGLDIHFIHVKPPQLPAGHTPKPLLMVHGWPGSFYEFYKIIPLLTDPKNHGLSDEHVFEV
ICPSIPGYGFSEASSKKGFNSVATARIFYKLMLRLGFQEFYIQGGDWGSLICTNMAQLVP
SHVKGLHLNMALVLSNFSTLTLLLGQRFGRFLGLTERDVELLYPVKEKVFYSLMRESGYM
HIQCTKPDTVGSALNDSPVGLAAYILEKFSTWTNTEFRYLEDGGLERKFSLDDLLTNVML
YWTTGTIISSQRFYKENLGQGWMTQKHERMKVYVPTGFSAFPFELLHTPEKWVRFKYPKL
ISYSYMVRGGHFAAFEEPELLAQDIRKFLSVLERQ
|
| External Links |
|---|
| GenBank ID Protein
| Not Available |
| UniProtKB/Swiss-Prot ID
| P07099 |
| UniProtKB/Swiss-Prot Entry Name
| HYEP_HUMAN |
| PDB IDs
|
Not Available |
| GenBank Gene ID
| J03518 |
| GeneCard ID
| EPHX1 |
| GenAtlas ID
| EPHX1 |
| HGNC ID
| HGNC:3401 |
| References |
|---|
| General References
| Not Available |


